Disease classifications in medicine are increasingly transformed by enhanced knowledge of molecular foundations, especially where clinical manifestations are diverse and illness trajectories are multifarious. There are multiple examples where biological differentiation has resulted in classification of diseases with remarkably similar clinical presentations and pathology into distinct disorders (
1,
2). Statistical modeling of clinical and biomarker data sets can facilitate redefinition and reconceptualization of complex human diseases (
3,
4). More basic knowledge of neurobiological architecture can enhance treatment research and outcomes (
5,
6) and support development of treatments tailored for patients’ unique etiopathologies (
7).
Biological reformulations of disease have revolutionized many medical disciplines, but classification and treatment of brain diseases subsumed by psychiatry rely on clinical phenomenology, despite the call for alternatives (
8,
9). Even bipolar disorder with psychosis and schizophrenia, the two major and ostensibly distinct psychosis categories, do not “breed true” (
10,
11). There is overlap in susceptibility genes and phenotypes across bipolar disorder with psychosis and schizophrenia (
12–
14) and considerable similarity between different psychotic disorders on symptoms, illness course, cognition, psychophysiology, and neurobiology (
15–
26). Drug treatments for these conditions overlap extensively (
27). “Psychosis” could be a final endpoint for multiple psychotogenic etiologies, as “congestive heart failure” is a common endpoint of cardiac, renal, and pulmonary disorders, all of which are best ameliorated with distinct treatments (for example, see reference
28). A useful complementary approach may include the development of a more neuroscience-based classification of the psychoses (
29).
To evaluate this possibility, we recruited individuals manifesting psychosis, a neurobiologically heterogeneous target population with unknown and certainly diverse etiologies. We collected a large panel of biomarkers of known relevance to psychosis and functional brain activity. Multivariate analyses were used to partition neurobiologically distinct subgroups of psychosis cases independent of clinical phenomenology. We refined a subset of the biomarker panel that differentiated people with psychosis from healthy persons, and we used those biomarkers to differentiate among (create distinct subgroups of) psychosis cases. The neurobiological uniqueness of the newly created psychosis categories was supported with meaningful external validators (for an illustration of the approach, see Figure S1 in the data supplement accompanying the online version of this article). Given the apparent distinctiveness of these subgroups, we call them psychosis “biotypes” (biologically distinctive phenotypes). Much like for other branches of medicine, the biotypes did not respect clinical phenomenological diagnoses (see references
30–
33). Identifying additional unique characteristics of these psychosis biotypes may facilitate novel clinical, basic, and molecular research (
34).
Discussion
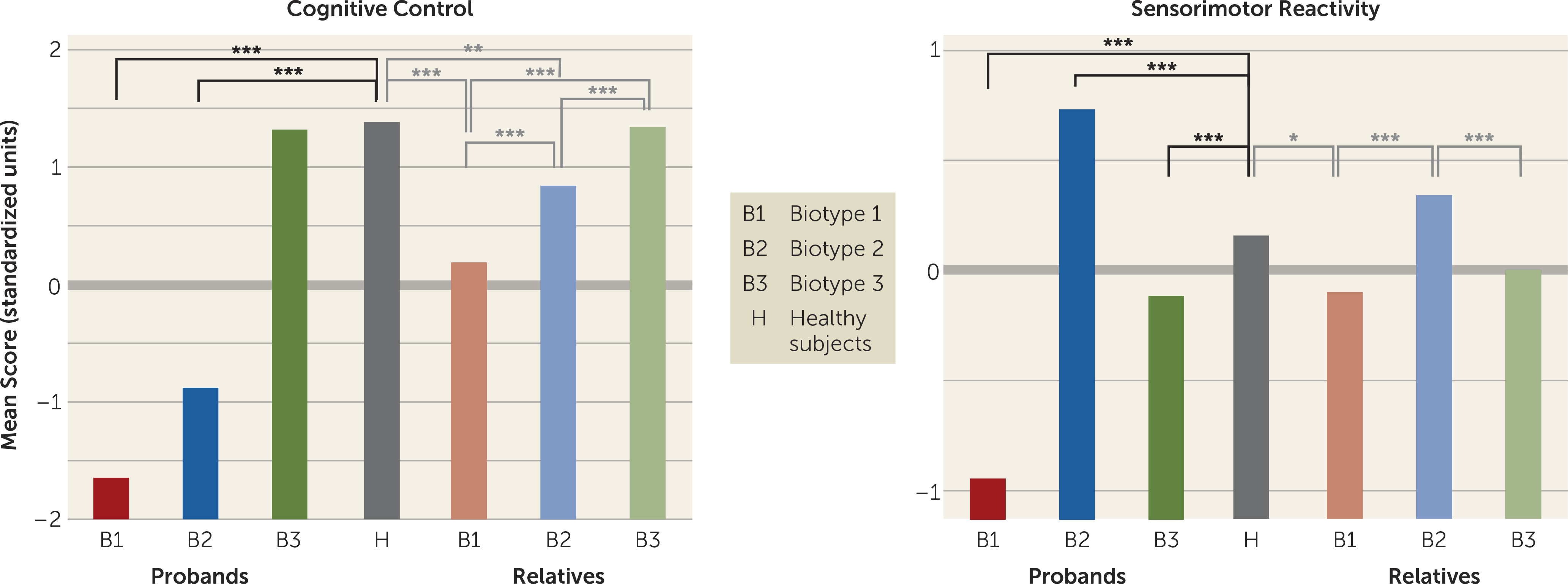
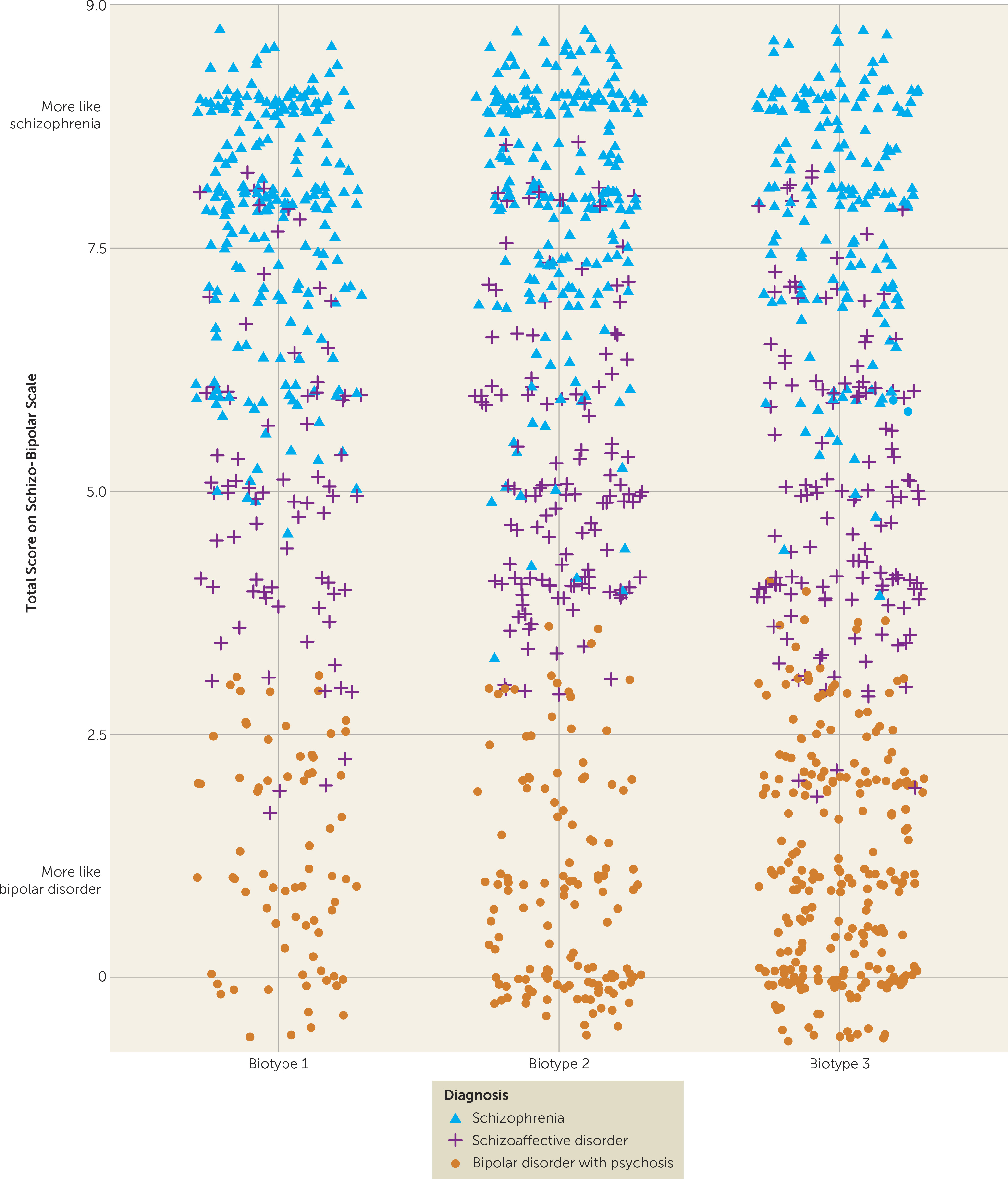
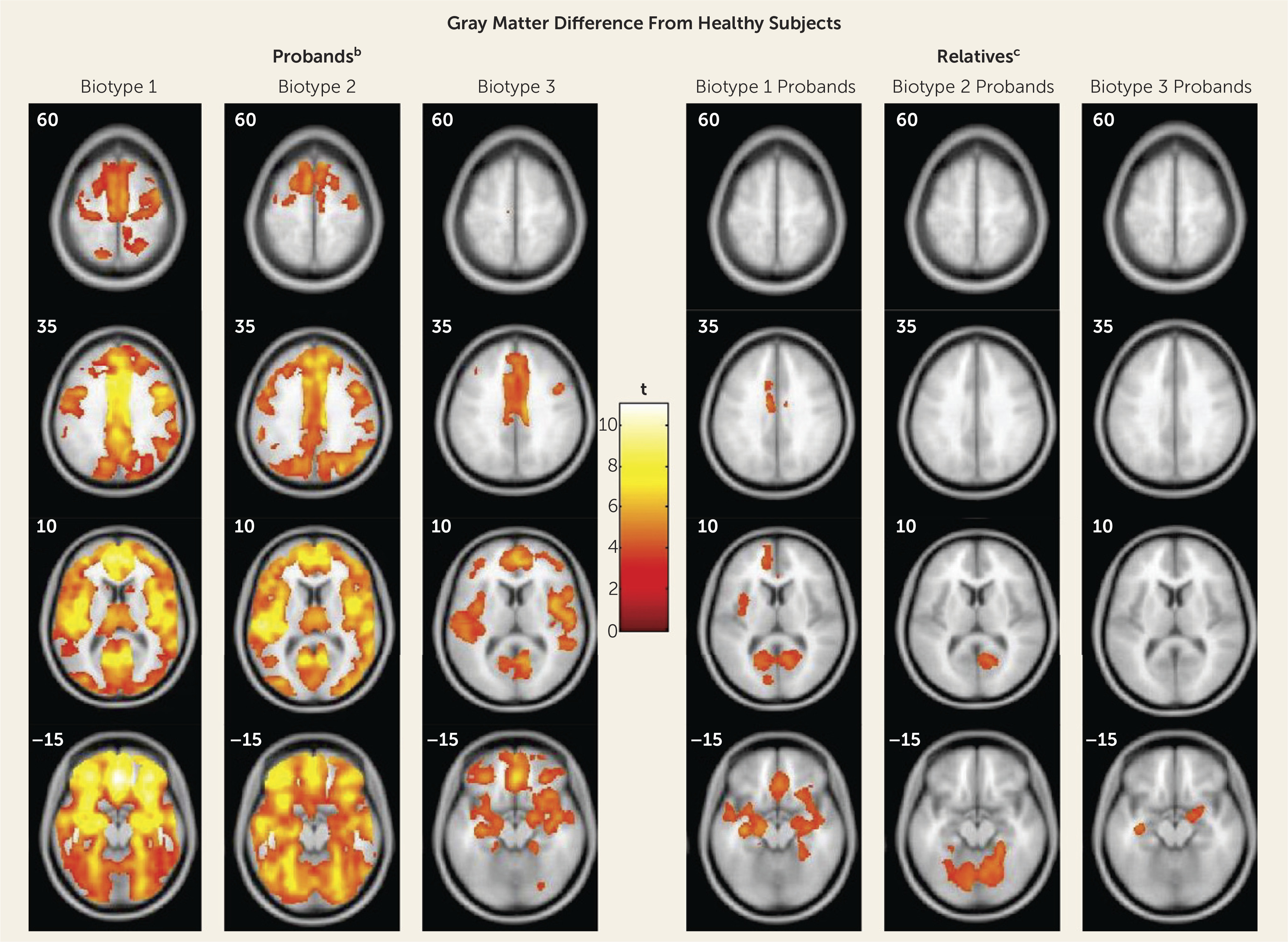
The neurobiological heterogeneity across the psychosis spectrum illustrates the difficulty with attempting to derive etiological and neurobiological distinctiveness from clinical phenomenology alone. The present approach drew on biomarker heterogeneity to identify brain-based psychosis biotypes independent of specific clinical features (other than presence of psychosis). Each biotype included all DSM psychosis categories, but probands diagnosed with schizophrenia were more numerous in biotype 1 (although 20% had bipolar disorder with psychosis) and probands diagnosed with bipolar disorder with psychosis were more numerous in biotype 3 (although 32% had schizophrenia), respectively. Measures not used in creation of the biotypes, including social functioning, brain structure, and characteristics of biological relatives, independently supported biotype distinctiveness. When considered across proband and relative data, the biotype subgroups were superior to DSM diagnostic classes in between-group separations on external validating measures, illustrating the former scheme’s superiority for capturing neurobiological distinctiveness.
An important feature of our approach was integration across numerous laboratory biomarker measures of psychosis-related neurobiological deviations. The statistically derived constructs, labeled “cognitive control” and “sensorimotor reactivity” given their constituents, have played prominent roles in previous attempts to describe neurobiological deviations in psychosis, most notably schizophrenia (
48). These multivariate constructs were superior to the individual biomarkers (had larger effect sizes) for distinguishing between subgroups. This was true for both biotype designations and DSM diagnostic classes. Epigenetics, etiological heterogeneity, pleiotropy, and variable expressivity, among other factors, influence phenotypic manifestations (
50), making it difficult for a single laboratory measure to be pathognomonic for complex psychiatric diseases.
These data also indicated that there may be multiple pathways to clinically similar psychosis manifestations. Biotype 1, the subgroup with the fewest probands, was most prototypical of the chronic, deteriorated, poor-outcome cases often considered to capture the essence of schizophrenia (
19,
23,
26,
29). These participants showed profound dysfunction on cognitive control and had severely compromised neural reactivity to even simple sensory events. Their first-degree biological relatives showed the same pattern of deviations. In contrast, individuals in biotype 2, although also demonstrating marked, but less severe, cognitive control dysfunction, were characterized by accentuated sensorimotor reactivity, a feature that has been previously described (
51). This pattern of deviations for biotype 2 probands was mirrored in their biological relatives. Nevertheless, biotypes 1 and 2 had highly similar clinical severity and evidence for familial psychosis risk. In contrast, individuals in biotype 3, the largest of the groups, showed less severe clinical psychosis manifestations, nearly normal cognition, and sensorimotor reactivity less distinguishable from normal.
These differential patterns of biomarkers across biotypes invoke an explanation for the marked heterogeneity in DSM diagnoses that is routinely observed across research laboratories, even on the same biomarker variables. Individuals with biotype 1, being the most compromised, might be more likely to be recruited in inpatient settings, while those with biotype 2 or 3 might be more prominent in outpatient clinics. In addition, researchers working in settings that captured mostly biotype 3 probands would justly conclude that studying neurobiological risk indicators (endophenotypes) for psychosis among biological relatives was a futile enterprise. Investigators in molecular genetic studies sampling high concentrations of biotype 3 probands also might conclude that a large proportion of the variance in psychosis genetic risk was captured by spontaneous mutations (
12–
14). Recruitment strategy and subject sampling, therefore, might influence determinations of beliefs concerning the core neurobiological features of psychosis.
These divergent biomarker patterns across biotypes also illustrate the difficulty with using solely clinical psychosis diagnoses as the “gold standard” for capturing neurobiological distinctiveness (
32,
33). The psychosis probands (as a group) showed at least some degree of cognitive control deviation, but the remarkable difference between diminution and accentuation of sensorimotor reactivity across individuals with psychosis would certainly lead to devaluing this group of measures for understanding psychosis neurobiology in mixed samples (the overall mean would be close to that seen among healthy persons). In contrast, molecular, pharmacological, and genetic studies directly comparing biotype 1, 2, and 3 subgroups, as defined by both cognitive control and sensorimotor reactivity constructs, could be useful for disentangling at least part of the etiological and pathophysiological heterogeneity purported to typify clinical psychosis (
52).
The biotype outcome provides proof of concept that structural and functional brain biomarker measures can sort individuals with psychosis into groups that are neurobiologically distinctive and appear biologically meaningful. These outcomes inspire specific theories that could be fruitfully investigated. First, biotypes 1 and 2 should be of greater interest in familial genetic investigations, while perhaps biotype 3 would be more informative for explorations of environmental correlates of psychosis risk, spontaneous mutations, and/or epigenetic modifications. Second, treatments for biotype 1 would be naturally directed both to profoundly compromised cognitive control and to correcting reduced neural activations that compromise signal-to-noise ratios for discerning environmental stimulus relevance (manifest in EEG signals). Third, biotypes 1 and 2 could be explored for potassium and/or calcium channel alterations and channel-based therapies that may correct neuronal hypo- or hyperexcitability (
53). It is possible that biotypes and/or related neurobiological parsing approaches will contribute to defining biological correlates of psychosis. Whether these constructs become important in psychosis research will depend on their usefulness, i.e., on their ability to foster and support incisive molecular, genetic, and treatment distinctions.
The present approach to parsing neurobiological variance among the psychoses has limitations that we attempted to minimize but could not completely overcome in this single project. First, the probands were mostly medicated, chronically psychotic, and tested at one time point; we have yet to demonstrate the longitudinal stability of these subgroupings, though the biomarkers themselves may be stable traits. Second, there was no replication sample in this initial data analysis, even though collection of a larger replication sample is in process. We did, however, test the accuracy of the neurobiological classification by using a jackknife procedure, and the data on first-degree relatives provided additional and important support for the subgrouping scheme (these data were not used in biotype construction). Third, the included biomarkers were chosen on the basis of their previously demonstrated success (at the initiation of this project) for distinguishing psychotic disorder subjects (probands and relatives) from healthy persons (
38). It is unlikely that this demonstration of a promising means for capturing neurobiological distinctiveness in psychosis describes all relevant possibilities; the inclusion of additional biomarkers may be useful in this regard. Fourth, the estimation of biotype membership, at present, requires extensive biomarker assessments. A means for estimating such membership, however, through clinical examination would be an extremely useful contribution to our understanding of the psychoses.