Cognitive deficits are the most common source of disability following traumatic brain injury
(1). However, the factors influencing cognitive outcome are poorly understood. One can see very different cognitive and functional outcomes in individuals with similar pre-injury intellectual/educational backgrounds and degrees of injury, suggesting that individual differences, including genotype, play important roles.
Because dopaminergic tone helps modulate memory, attention, and frontal-executive functions in traumatic brain injury
(2), genes influencing central dopaminergic function are attractive candidates for study. The gene for the dopamine D2 receptor (DRD2) has several common polymorphisms. One of these, the “TaqI A” polymorphism, is a C/T single nucleotide polymorphism (SNP) about 10 kb centromeric to the stop codon of DRD2 on 11q23 (SNP rs1800497). The C allele results in a TaqI restriction site; the T allele does not. In the older literature, the T allele was described as the A1 allele. Consistent with the newer convention, we will refer to this as the T allele. The presence of the T allele is associated with a 40% reduction in the expression of D2 receptors in the striatum and perhaps other cortical regions, without change in receptor affinity
(3,
4). This association is most likely a result of linkage disequilibrium between this polymorphism and a functional DRD2 polymorphism, since rs1800497 itself is a nonsynonymous coding polymorphism in the adjacent but functionally unrelated gene ANKK1. The alternative hypothesis that ANKK1 regulates DRD2 levels has not been ruled out, however
(5). We hypothesized that individuals with the T allele would have poorer cognitive function following mild traumatic brain injury.
Method
Consecutive patients who met American Congress of Rehabilitative Medicine criteria for mild traumatic brain injury
(6) were recruited from a level-1 trauma center emergency department. Healthy comparison subjects were recruited through advertisements. Exclusion criteria included history of neurological disorder, previous traumatic brain injury with clear loss of consciousness, substantial systemic medical illness, or current DSM-IV axis I psychiatric diagnosis (based on the Structured Clinical Interview for DSM-IV
[7]). The study protocol and informed consent were approved by the Dartmouth College Committee for the Protection of Human Subjects. Written informed consent was obtained from all participating subjects.
Participants were given the Wide-Range Achievement Test, 3rd ed. (WRAT-3) reading subtest
(8), the WAIS-III Block Design subtest
(9), the California Verbal Learning Test
(10,
11), and the Continuous Performance Test
(12).
QIAGEN’s blood mini kit (QIAGEN, Alameda, Calif.) isolated DNA from peripheral blood samples. The genotype was determined by using real-time polymerase chain reaction and Eclipse MGB probes (Epoch Biosciences, Bothell, Wash.) as described in the SNP500 database (http://snp500cancer.nci.nih.gov/snp.cfm).
Between-group differences in demographic characteristics were examined by t test. Group differences for neuropsychological measures were analyzed by analysis of variance with diagnosis, allele type, and the interaction included in the model. Bonferroni adjustment was used to determine significant between-group differences. All tests used two-tailed comparisons (p<0.05).
Results
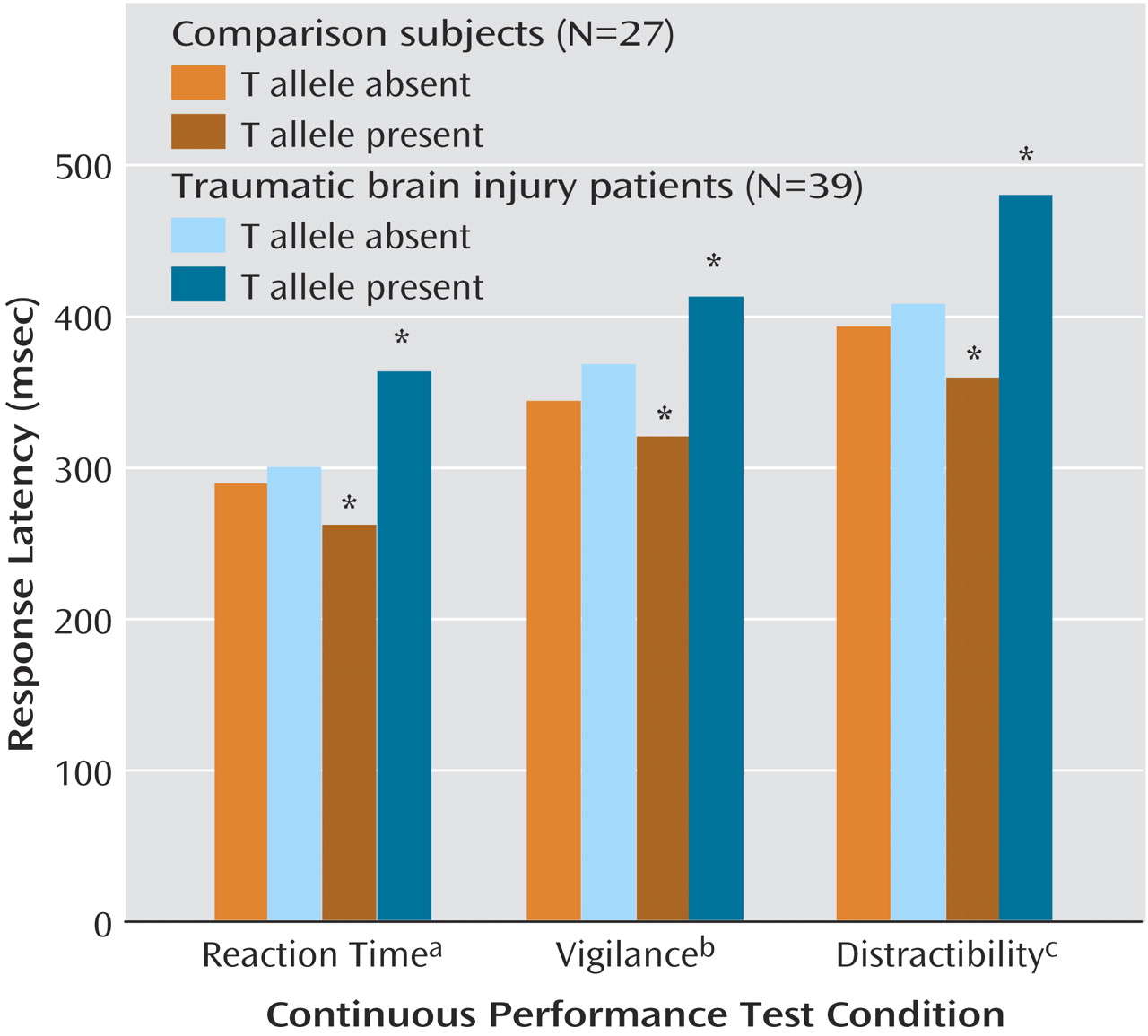
Thirty-nine individuals with mild traumatic brain injury and 27 healthy comparison subjects were studied. The subjects with mild traumatic brain injury were studied a mean of 38.4 days (SD=24.4) after injury. The mean age of the subjects with mild traumatic brain injury was 31.8 years (SD=13.2), and the mean age of the comparison subjects was 32.0 (SD=9.8). Over 99% of both groups were Caucasians of European descent. There were no significant between-group differences in gender (21 men and 18 women in the brain injury group and 11 men and 16 women in the comparison group), years of education (mean=14.2, SD=2.6, for the subjects with mild traumatic brain injury and mean=15, SD=2, for the comparison subjects), or parental education (mean=14.1, SD=2.5, for the brain injury group and mean=14.2, SD=2.6, for the comparison group). There were also no between-group differences in baseline intellectual function according to WRAT-3 reading score (mean=104.9, SD=9.9, for the subjects with mild traumatic brain injury and mean=109.3, SD=7.5, for the comparison subjects) and WAIS-III Block Design raw score (mean=29.2, SD=22.8, for the subjects with mild traumatic brain injury and mean=27.9, SD=17.9, for the comparison subjects). The distribution of T allele was identical in the two groups (11 subjects with mild traumatic brain injury and 11 comparison subjects).
In the combined group of patients with mild traumatic brain injury and comparison subjects there were 44 C/C, 22 C/T, and no T/T participants. The T-allele-positive participants with mild traumatic brain injury did not differ from the T-allele-negative participants with mild traumatic brain injury in injury severity (Glasgow Coma Scale
[13] score, estimated unconsciousness, duration of posttraumatic amnesia). There was a significant main effect of allele on recognition performance on the California Verbal Learning Test: lower performance was associated with presence of the T allele (T allele present: mean=13.6, SD=3.7, versus T allele absent: mean=14.9, SD=1.6) (F=4.10, df=1, 60, p<0.05). There was a significant main effect of diagnosis (mild traumatic brain injury versus comparison) on Continuous Performance Test response latency measures (simple reaction time [F=9.01, df=1, 61, p=0.004], vigilance [F=8.68, df=1, 60, p=0.005], and distractibility [F=8.52, df=1, 61, p=0.005]). There was also a significant diagnosis-by-allele (T-allele-positive versus T-allele-negative) interaction for each Continuous Performance Test condition (
Figure 1). The significance of the interaction was largely attributable to slower response latencies in the T-allele-positive subjects with mild traumatic brain injury than in the T-allele-positive comparison subjects, as well as a lack of significant differences between the other groups. T-allele-positive and T-allele-negative comparison subjects did not differ significantly in any of the Continuous Performance Test conditions.
Conclusions
These preliminary results suggest that polymorphisms having an impact on central dopaminergic function can influence cognitive performance and that the impact differs in subjects with mild traumatic brain injury and healthy comparison subjects. When both groups were combined, T-allele-positive individuals performed more poorly on the California Verbal Learning Test recognition task than T-allele-negative individuals. T allele status also affected response latencies: T-allele-positive subjects with mild traumatic brain injury performed most slowly on all Continuous Performance Test measures. In our previous research
(14), response latencies have been a consistent area of neuropsychological deficit within the first month after mild traumatic brain injury. We were surprised that the T-allele-positive comparison subjects had the lowest response latencies. This may be a result of a relatively small number of subjects, because the T-allele-positive and T-allele-negative comparison subjects did not differ significantly from each other. Several factors could be affecting response latencies, including subtle changes in attention, perceptual motor processing, or speed of information processing. Different cognitive domains may respond differently to changes in central dopaminergic tone, and we have presented evidence that a recent mild traumatic brain injury alters this response
(15). Further work is needed to clarify the relative contributions of these different factors.
The modest effect size of the T allele on the California Verbal Learning Test (0.21) is consistent with other reported allelic effect sizes on cognitive function and other polygenic traits
(16). We cannot rule out the possibility that the observed effects may be due to another allele in linkage disequilibrium with the T allele. Stratification effects in this study group are unlikely because the T-allele-positive and T-allele-negative groups had virtually identical racial/ethnic populations. These findings need replication in a larger group of subjects.