Specific copy number variants (CNVs) have been robustly associated with intellectual disability, autism, and schizophrenia. Most of the focus in the literature has been on documenting the existence of these phenomena. There are few data to guide therapeutic choices for these “orphan” diseases. Here, we call for systematic and longitudinal case reports that, if carefully conducted, may provide crucial initial knowledge to guide therapeutic interventions. We provide a step-by-step overview, a tailored set of consensus criteria for high-quality case reports, and a specific set of learning resources.
Imagine an initial psychiatric interview that begins with the chief complaint, “I have a de novo 16p11 duplication, and they say I have schizophrenia and Asperger’s. What does this mean for me and my family? How can you help me?” Provided that the patient’s report can be verified, this chief complaint has a strong scientific and empirical basis. This CNV results in changes in the number of copies of approximately 30 genes located on a 600,000 base pair region on the short arm of chromosome 16, and this rare genetic mutation increases risk for autism and schizophrenia (see Supplemental Note 1 in the online supplement). How exactly would you help? What evidence would support your clinical choices? On the basis of current knowledge, a clinician should explore the possibility of other psychiatric diagnoses (e.g., major depression, and speech and language delay), be alert for the presence of general medical abnormalities, including renal and urinary malformations, and consider the need for genetic counseling. However, while there may be broader implications for the patient and family, to the best of our knowledge, there are few relevant data that inform the psychiatric management of this patient. Thus, how can our field cooperate to rapidly increase knowledge relevant to clinical management?
Although this scenario might seem far-fetched, genetic evaluations are increasingly part of the standard of care in psychiatry. A “genetic workup” is increasingly part of the clinical evaluation of children with moderate to severe intellectual disability, marked developmental delay, and autism, and it is justifiable for adults who present similarly or who have complex presentation (see below). In fact, psychiatrists who treat individuals with severe psychiatric disorders (including ourselves) are certain to have encountered patients with important genetic changes—and we probably did not know it. The rapid pace of progress in medical genomics means that these topics and their implications will be unfamiliar to many clinicians, and a number of educational resources are available (see Table S1 in the online supplement).
Normally, children inherit a paternal and a maternal copy of every autosome (chromosomes 1–22). Occasionally, there are errors in the meiotic or mitotic machinery so that large regions, often containing multiple genes, are lost or gained. Such changes are termed CNVs. These can occur at the level of a whole chromosome (e.g., trisomy 21, 48.1 megabases) or at finer levels (hundreds of kilobases or smaller, as with the 16p11 CNV). Many pathogenic CNVs recur because of regional genomic features and can be found worldwide (
1). Genomic studies have established the etiological importance of specific CNVs for psychiatric outcomes, with most of these CNVs associated with variable outcomes (pleiotropy), including moderate to severe intellectual disability, developmental delay, autism, tics, dyscoordination, and schizophrenia (
Figure 1).
In samples seen in clinical psychiatry, current estimates suggest that a clinically or etiologically relevant CNV is likely to be present in around 2%−3% of people with schizophrenia, 10% of people with autism, and 25% or more of people with intellectual disability (
2–
4). In general, the greater the severity, the higher the prevalence (i.e., lesser in unselected population surveys, and higher in the most severely ill).
The accumulation of disease-relevant genomic data begs the question of how this should affect clinical decision making, but the clinical knowledge base is limited. We need more treatment and management data to guide therapeutic choices for people with a pathogenic CNV and a severe psychiatric disorder, and to be able to address questions about familial risk. Although the associations of specific CNVs as etiological risk factors are secure, we do not now have an adequate knowledge base to inform the practice of clinical psychiatry. In almost all instances, the emphasis is on diagnostic features, unusual presentations, and information particularly salient to clinical geneticists, neurologists, and pediatricians. In reviewing the literature, we found that the clinical knowledge base relevant to psychiatric management is sparse—there are few data relevant to the question, “How can you help me?”
Although as a group these CNVs are a relatively common etiological risk factor, the individual CNVs are rare and are almost all “orphan diseases” (i.e., diseases affecting less than 200,000 people in the United States, equivalent to a lifetime prevalence <0.06%; see Supplemental Note 2 in the
online supplement). Accruing sizable samples for systematic study requires international consortia, considerable expense, and many years of effort. A prime example is the International Consortium on Brain and Behavior in 22q11.2 Deletion Syndrome (22q11DS) (
5). This consortium has assembled a sample of 1,616 psychiatrically well-characterized cases and obtained genomic data on >300 adults split evenly into those with schizophrenia and those without. The goals of this study are to understand how other genetic and nongenetic factors influence the expression of schizophrenia that may be of wider relevance to the general population, to provide information on the precursors and antecedents of schizophrenia, and to serve as a base for future longitudinal studies aiming to study the neurodevelopmental trajectories of deletion carriers. The outcomes of the study are beginning to appear and include the largest characterization of psychiatric outcomes in 22q11DS to date and the identification of childhood antecedents of psychotic outcomes (
6,
7).
It is reasonable to ask whether improved therapeutics are likely. We do not know. However, as a proof of concept, Deborah Levy and colleagues recently reported two individuals with psychotic disorders and very rare CNV triplications of the glycine decarboxylase gene (
8). Under the assumption that
N-methyl-
d-aspartate hypofunction was present because of increased glycine catabolism (and low levels of brain glycine and
d-serine), the authors demonstrated clinical improvement in psychotic and mood symptoms with oral glycine supplementation (in a double-blind placebo-controlled study). If a clinician were to encounter a patient with this CNV in the future, this report would provide reasonable therapeutic guidance.
We propose a systematic effort to obtain clinical data useful to management in clinical psychiatry. We effectively propose “clinical crowdsourcing,” which combines the advantages of a distributed effort with a comprehensive and systematic structure to yield high-quality case report and case series knowledge to inform clinical psychiatric management. Our proposal has the following steps:
1) Detection. We need to test more patients for CNVs. In fact, there is a strong case for universal testing for some psychiatric disorders, particularly for early-onset and severely impairing conditions (moderate to severe intellectual disability and autism), as well as chronic psychotic disorders in adults. A positive result is relevant to clinical management—most large CNVs are multisystem disorders associated with increased risk of cardiac, neurological, endocrine, renal, hematological, and digestive complications (
9). We encountered a patient whose idiopathic thrombocytopenia was chased after for years but was almost certainly a consequence of 22q11DS (
10). We know of drug companies working on therapeutics for specific CNVs; if targeted medications become available in the future, we need to know the patients for whom these therapies may be indicated. The presence of a CNV can be relevant to reproductive planning, but the wide range of psychiatric and cognitive outcomes and incomplete penetrance call for nuanced and informed genetic counseling (
11).
CNV testing is typically offered for children with intellectual disability, developmental delay, or neurodevelopmental disorders. But CNVs are also risk factors for adult psychiatric cases, particularly schizophrenia. What clinical features increase the likelihood of the presence of a CNV and should act as “flags” for targeted testing in adult psychiatry? We are not aware of systematic studies investigating such clinical features, and there is clearly a pressing need for these, as we argue below. However, based on current knowledge, we would highlight premorbid low intelligence, a history of childhood-onset neurodevelopmental disorder, congenital malformations, dysmorphic features, and developmental delay (missing developmental milestones). A family history of schizophrenia or other neurodevelopmental disorders may also be relevant (although pathogenic CNVs frequently occur de novo, so a family history in a parent is often absent). Finally, we never discount the importance of the intuition of experienced clinicians, or the sense that a particular patient is distinctively different from others in the same diagnostic category. Examples here include very severe symptoms or extreme treatment refractoriness and prominent general medical comorbidity.
Exactly what test to order depends on the clinical context and on the availability and cost of technologies. We offer the following general considerations. First, consultation with a clinical geneticist and/or a genetic counselor is usually important. Second, for adult psychotic disorders, evaluation of CNVs would be a typical starting point. One technology for this purpose is chromosomal microarray, which can identify the presence of large pathogenic CNVs (and costs approximately $300). Third, for early-onset, severe psychiatric disorders, a typical panel would include chromosomal microarray and resequencing of the protein-coding portion of the genome (whole exome sequencing) or whole genome sequencing. Applying the same technologies to both biological parents can help in prioritizing detected variants and in determining whether a variant is de novo or inherited.
2) Capture the needed data. In most instances, case reports that describe only the co-occurrence of a known CNV with a psychiatric disorder may not be particularly notable for associations that have been extensively documented. At the same time, there may be some novel or remarkable feature that would support publication (e.g., our report of a man with 22q11DS and Huntington’s disease [
10]).
In our opinion, we need case reports and case series that have two key features: a comprehensive initial assessment, and systematic description of the longitudinal course and the impact of therapeutic efforts.
Initial assessment should include a multi-informant history of salient events in pregnancy, birth, childhood development, adolescence, and adulthood. Assessments or indications of intellectual function across development are very important (if not essential). Collecting such data from a psychiatric perspective is generally routine in clinical practice. However, for CNVs, there should be particular attention on congenital and multisystem abnormalities across all organ systems. The presence of general medical comorbidities should be sought, perhaps in collaboration with medical colleagues from other relevant specialties, bearing in mind those conditions known to be associated with a specific CNV. Finally, a thorough family history should be obtained with a focus on the range of neurodevelopmental disorders, including schizophrenia, that are associated with risk CNVs. Many would suggest that brain imaging using MRI is important.
The critical missing ingredients are the longitudinal course and impact of therapeutics. There needs to be a systematic description of age-dated therapeutics across all modalities (pharmacological as well as behavioral and psychological). These also need to be connected to age-dated assessments of functional capacity, occupation, and role function, as well as inpatient, emergency, and outpatient treatment. These data should be combined to establish correlations as to what therapeutic strategies were optimal for this particular patient. In effect, this would be a variant of the “N of one” clinical study.
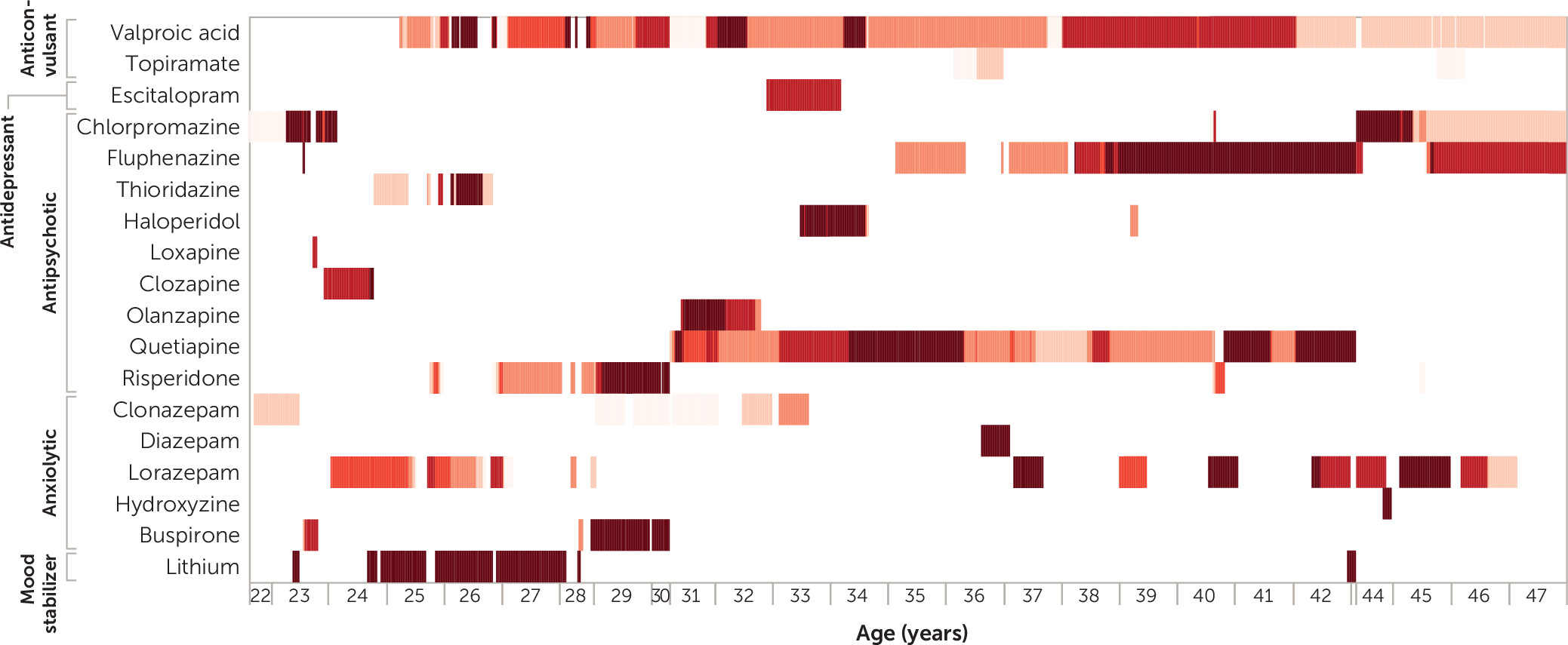
We note that modern data science (
12) has many excellent tools for obtaining, refining, summarizing, and presenting complex longitudinal data. This is increasingly easy to accomplish given the availability of electronic medical records. For example,
Figure 2 took 5 minutes to make but captures 25 years of pharmacotherapy for a person with highly treatment-resistant psychosis.
3) Publish a case report. It is then important to let the scientific community know what you have learned. The particular focus should be on what worked, what did not work, and what you might do differently if you could do it over. We strongly advocate for following explicit guidelines for the structure and content of a case report—for instance, the CARE criteria, which were developed in the general medical context to improve completeness and transparency and to facilitate the systematic aggregation of information across reports (
13) (see Table S3 in the
online supplement). In particular, the title should include “case report” and standard terms for the specific genetic change and the psychiatric diagnosis. Reasonable examples of CNV case reports and case series may be found in references
10 and
14. There are multiple target journals for these case reports. Particularly detailed or notable case reports may appear in higher profile journals; however, it is critical for these case reports to be findable via inclusion in searchable resources like PubMed and PubMed Central. There are multiple open-access journals that are devoted to case reports and that are indexed in PubMed. A basic web search for “case report journals” found 10, and there are even overviews on the choice of case report journal (
15).
The potential benefits of what we propose are to allow other clinicians to benefit from the experiences of colleagues in the struggle to deliver effective clinical management and to identify treatment options for individuals with rare pathogenic CNVs and severe mental illness. Many patients with CNVs experience protracted and stressful “diagnostic odysseys” in referrals to multiple specialists for organ-specific evaluations when, in the end, the root cause is a CNV with effects on multiple systems. Minimizing time to CNV identification would minimize such odysseys and rapidly signpost potentially appropriate nonpsychiatric assessments. Moreover, it is possible that detailed evaluation of these rare patients could yield therapeutic and etiological ideas relevant to patients with idiopathic forms of these disorders.
“Bespoke therapeutics” may ultimately be an important benefit. For certain rare CNVs, the literature may suggest a therapy that is uniquely tailored to an individual with a particular CNV. Current examples include the use of oral glycine in CNV triplications of the glycine decarboxylase gene (
8) and the anecdotal use of oral magnesium supplementation in Burnside-Butler syndrome (a 15q11.2 CNV deletion that affects
NIPA1 and
NIPA2, which are involved in brain magnesium transport) (
16). We contend that by rapidly sharing and disseminating clinical and therapeutic findings, we may be able to build on these small but important beginnings.
A focus on copy number variation can also improve diagnostic classification. Many CNVs have highly variable clinical presentations that can include combinations of intellectual disability, specific learning impairments, autism, attention deficit hyperactivity disorder, anxiety and mood disorders, and psychotic disorders. In current diagnostic schemas, these are coded according to the clinical presentation; however, while this has value, it is crudely akin to coding rash, fever, headache, photophobia, and altered mental status instead of N. meningitides meningitis. Given the range of psychiatric disorders and medical comorbidities associated with these CNVs, a primary diagnosis that includes reference to the CNV may be more parsimonious to capture and may alert clinicians to the full range of important sequelae.
Several challenges are noted above, particularly the greater need for longitudinal and process outcome data (as opposed to merely documenting the co-occurrence of a CNV and a clinical presentation). An additional challenge is that advances in psychiatric and medical genetics mean that psychiatrists (particularly those in training) will need to understand how to generate, interpret, and explain genetic findings to their patients as well as how to use these data clinically (
17). There are multiple ways to obtain direct-to-consumer genomics (many of dubious clinical utility), and clinicians will increasingly be faced with questions about their relevance. For interested readers, Table S1 in the
online supplement contains a list of learning resources. There is clearly a need to embed genetics training deeply in residency training programs and to upskill practicing psychiatrists. Nurnberger et al. provide recommendations for psychiatry residency training and note that “the basic principles of genetics … are essential to current psychiatric patient care” (
18).
A key challenge is clinical synthesis. For example, for the patient introduced at the start of this commentary, how would a clinician efficiently, effectively, and accurately extract clinical guidance from the literature? Literature reviews are usually a great starting point (if they exist, but with the caveat that they miss case reports since submission). Moreover, they may not cover the exact clinical need. We suggest there is an unmet need: we need a structured and curated online database that captures case report data at the interface of genetics and clinical psychiatry. This is largely informatics but requires a funder to champion and support the idea. The idea is straightforward: to systematically capture the genetic mutation, the clinical phenotypes using a structured vocabulary (e.g., the Human Phenotype Ontology), the therapeutics attempted, and the therapeutic outcomes and adverse events. A reasonable model for this is the DECIPHER database in the United Kingdom (
https://decipher.sanger.ac.uk). In addition, such a database could serve as the basis for research and grant applications to derive clinical and biological hypotheses as well as to support accrual of “orphan” patients for future systematic studies.
The existence of a sizable body of case reports—particularly if prepared to a high standard—would provide practical guidance for the clinical psychiatric management of people with a psychiatric disorder and a pathogenic CNV. If we were to have 10 such reports for the vignette at the start of this essay (for a de novo 16p11 CNV and putative diagnoses of schizophrenia and Asperger’s syndrome; “how can you help me?”), one could synthesize the reports to derive an empirical management plan. One might also discover a psychiatric colleague with particular expertise in treating adult patients with 16p11 CNVs, and a conversation or e-mail exchange could be helpful.
Better case report data are better than nothing. However, ultimately, there is a need for more adequately powered studies that are able to relate specific genomic risk factors to clinical and neurocognitive outcomes and therapeutics. The individual CNVs are rare, but the clinical psychiatric phenotypes with which they are associated are not. The variable expressivity and pleiotropy seen offer an important opportunity to investigate the role of other genetic and environmental factors in modifying psychiatric outcomes, to deliver findings of relevance to psychiatric disorders more generally, to study groups of subjects in early childhood at high risk of later childhood and adult disorders, and to study the developmental course of psychiatric disorders and identify potentially modifiable antecedents and modifiers. It is gratifying that the National Institute of Mental Health has identified the potential of these studies and the need to assemble coordinated multidisciplinary and multisite teams capable of combining genomic data with comprehensive dimensional and categorical phenotype data. This is an excellent beginning, but we believe that more attention to and funding for this emerging area are required.
Acknowledgments
The authors are indebted to Drs. Rick Josiassen, Maya Lichtenstein, Marty Farrell, and Robert Stowe for helpful discussions.