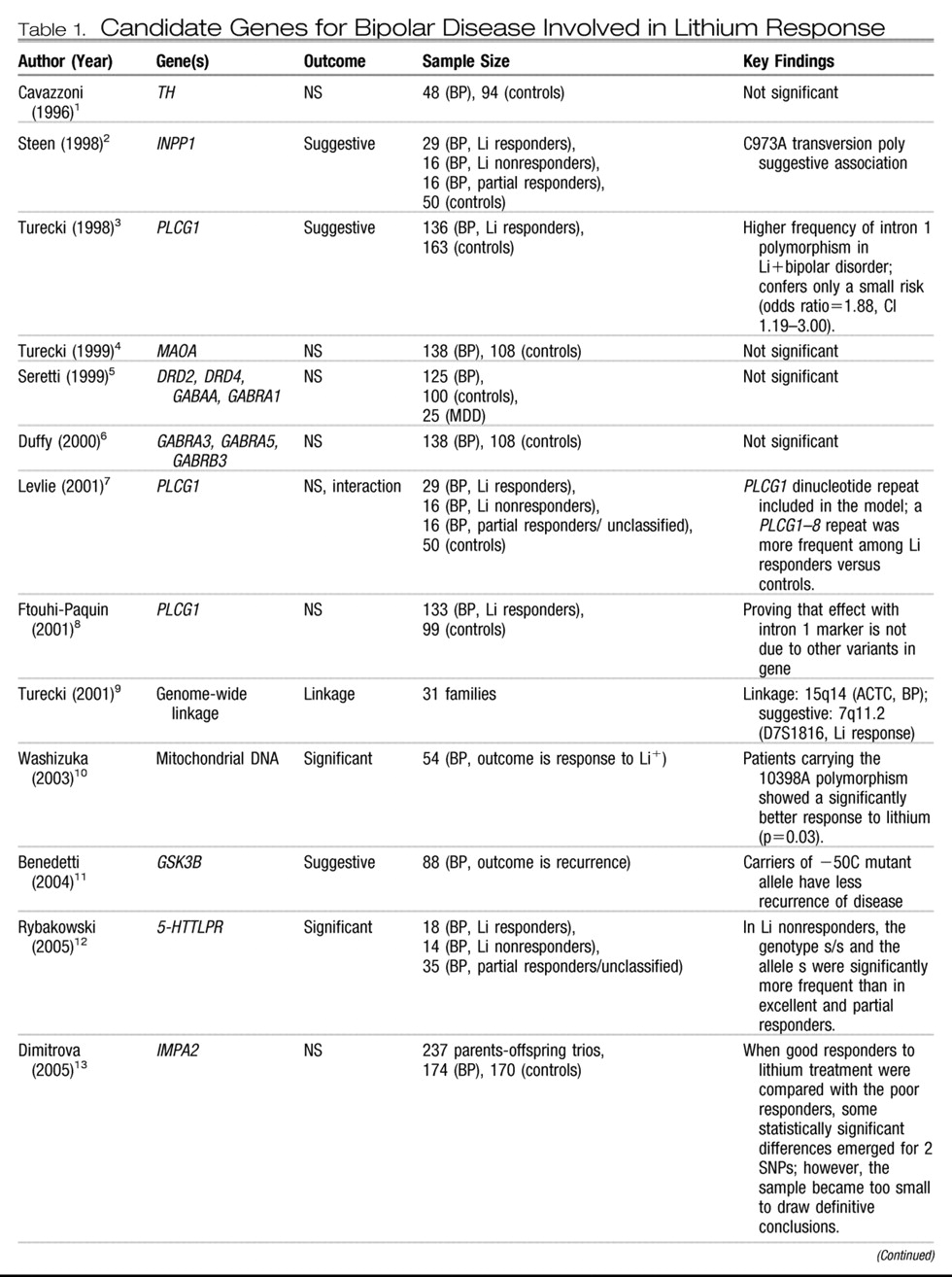
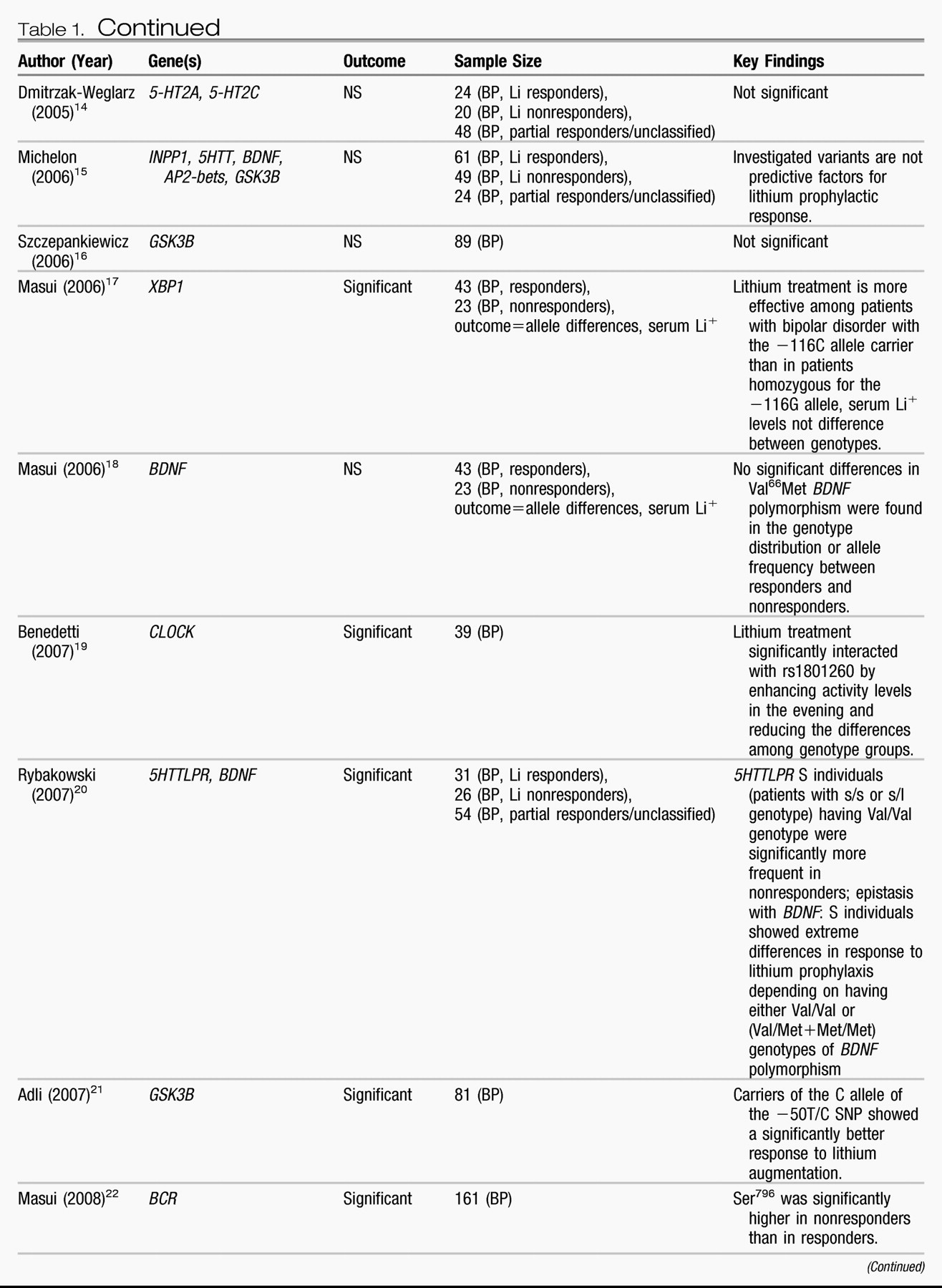
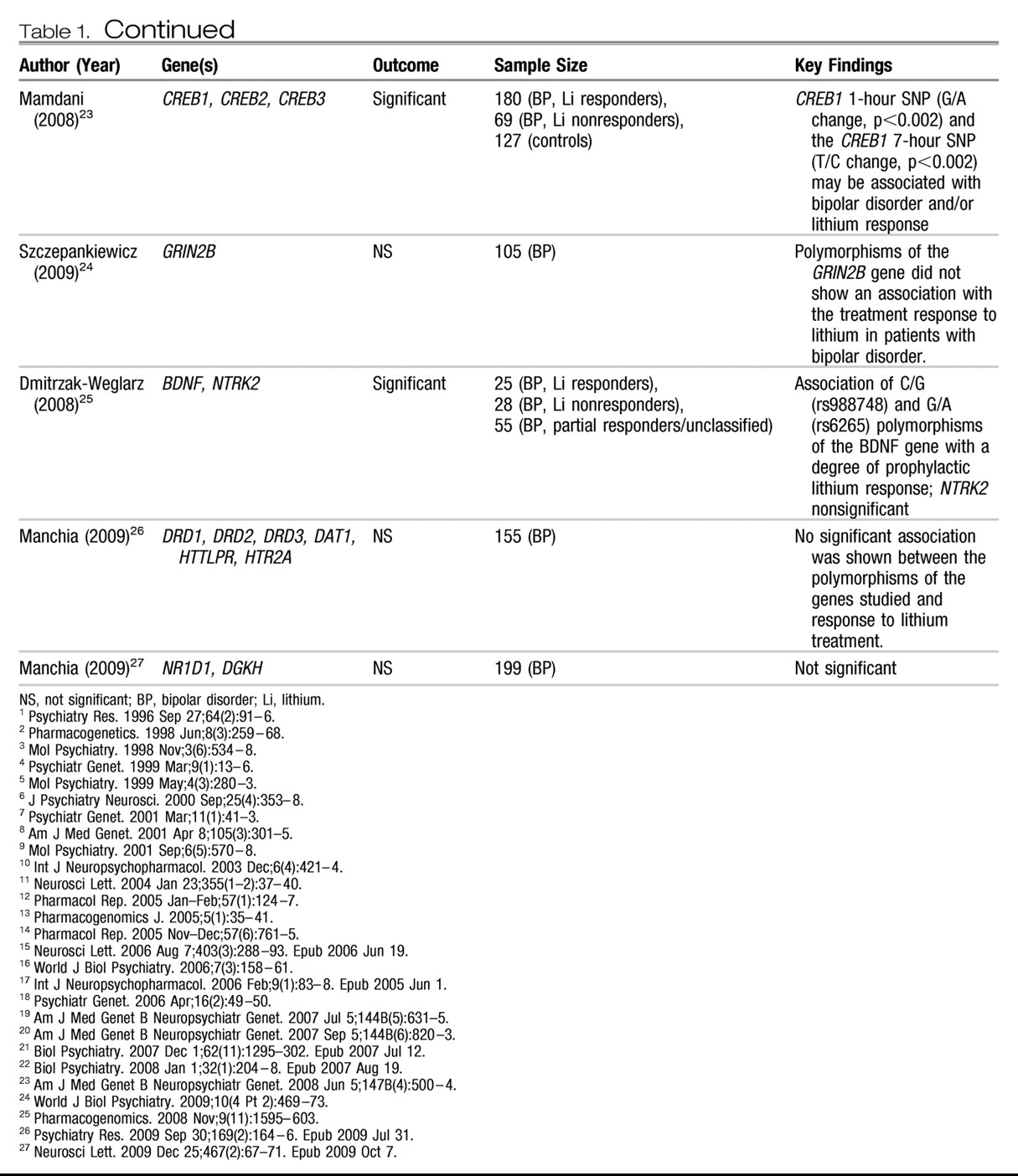
Candidate genes.
Meta-analysis of five twin studies has demonstrated that the heritability of major depressive disorder (MDD) is estimated at 37%, suggesting that there are both environmental and genetic factors influencing risk for MDD (
4). One of the most well-studied neural systems in MDD is the serotonergic pathway. The first reported associations were with a variable number tandem repeats (VNTR) variant and an insertion/deletion polymorphism near the serotonin transporter gene (
5,
6). The
5-HTT-linked insertion/deletion polymorphic region (5-HTTLPR) maps 1.4 kilobases upstream of the translational start site. The short variant has been shown to restrict transcriptional activity of the serotonin transporter (
5-HTT) promoter, leading to low functional expression of the
5-HTT; thus, the risk allele is most likely acting via changing expression of the
5-HTT transcript, resulting in less presynaptic uptake of serotonin (
6,
7). Further work has focused on environmental influences modifying the relationship between 5-HTTLPR and risk. Caspi et al. (
8) demonstrated that individuals with one or two copies of the short allele of the 5-HTTLPR promoter polymorphism exhibited more depressive symptoms, diagnosable depression, and suicidality in relation to stressful life events in children (i.e., abuse and neglect) than individuals homozygous for the long allele. Grabe et al. (
9) found significant interactions between genotype, unemployment, and chronic diseases in females but not in males, further validating the modifying response of trauma and
5-HTT variation. This result further implicated the short allele of the 5-HTTLPR promoter polymorphism in vulnerability to social stressors and chronic disease.
In terms of pharmacogenetic response, serotonin reuptake inhibitors (SSRIs) act via inhibition of presynaptic serotonin reuptake; therefore, it makes sense to evaluate
5-HTT genotypes and SSRI response. A recent study comparing both the 5-HTTLPR promoter polymorphism and an intronic VNTR in
5-HTT in a group of SSRI nonresponders with a referent sample found that women with the short allele of 5-HTTLPR were less likely to respond to SSRI treatment, whereas the different alleles of the intronic polymorphism had no affect on response (
10). This result was in contrast with results from a study on 241 Korean inpatients examining
5-HTT genotypes and fluoxetine or sertraline response (
11). In the Korean patients, the short allele of 5-HTTLPR was associated with a positive response to treatment. This result reflects different outcomes due to either ethnicity effects or differences in study design. Another possibility is that the disparate results are indicative of
5-HTT being a false-positive response, which is supported by the negative findings from the STAR*D trial (
12). Further work needs to be performed to assess
5-HTT and SSRI response.
Additional work on serotonergic pathways and MDD has implicated the serotonin 5-HT2A receptor (
HTR2A) and more recently the serotonin 5-HT3A receptor (
HTR3A). Whereas most of the studies of serotonergic pathways have focused on schizophrenia and psychosis (see next section), McMahon et al. (
13) found an association between variation in intron 2 of
HTR2A and response to citalopram in patients with MDD. Citalopram down-regulates the serotonin 2A receptor, which is encoded by the
HTR2A gene. Another important factor in this association was that the allele involved in citalopram response was more frequent in Caucasian subjects, and treatment effectiveness is reduced in African American subjects. Therefore, the
HTR2A allele might account for some of the ethnic differences in treatment response in MDD. Another study examining the −42C>T of
HTR3A demonstrated that CC carriers had frontolimbic gray matter alterations, which were potentiated by early life stress and these changes could increase risk for MDD (
14).
Besides serotonin, the other major pathway that is crucial in the causality as well as the treatment of depression is the hypothalamic-pituitary-adrenal (HPA) axis. The HPA axis is a major part of the neuroendocrine system that controls the reaction to stress as well as many other bodily functions. There is a complex interaction between the different HPA axis thyroid hormones in MDD (
15), and recent evidence suggests that allelic variation is a factor in modifying both risk and drug response.
The first member of the HPA axis that was implicated in MDD was the FK506-binding protein 5 gene (
FKBP5). Binder et al. (
16) genotyped variants in eight genes involved in the HPA axis (
16) and found that variation in
FKBP5 was significantly associated with increased recurrence of depressive episodes as well as faster response to antidepressant drug treatment. The mapped risk variants were also associated with increased intracellular FKBP5 protein expression, which triggers adaptive changes in the glucocorticoid receptor and, thereby, modifies HPA axis regulation.
The second HPA axis gene to be associated with MDD was the corticotropin-releasing hormone receptor 1 (
CRHR1) gene. Corticotropin-releasing hormone is the principal neuroregulator of the HPA axis and plays an important role in coordinating the endocrine, autonomic, and behavioral responses to stress and immune challenge. The first associations, which mapped three variants, one of which was associated with disease risk, were found in Chinese cohorts (
17). That work was extended both within the same cohorts (
18) as well as in another cohort of Mexican Americans (
19), showing haplotype affects with treatment response for fluoxetine or desipramine. More recent work has now included
CRHR1 gene × trauma interactions on MDD risk, showing that certain alleles act synergistically with child abuse to increase risk (
20).
More recently, polymorphisms within the corticotropin-releasing hormone-binding protein (
CRHBP) gene were assessed with respect to response to citalopram using the STAR*D samples.
CRHBP is another member of the HPA axis and is a high-affinity binding protein that inactivates corticotropin-releasing factor. One variant (rs10473984) showed a significant association with both remission (and reduction in depressive symptoms in response to citalopram) (
21).
Along with serotonin, there is some evidence to suggest that norepinephrine pathways are involved in MDD and treatment response. Two studies have examined drug response and variation in the norepinephrine transporter gene (
SLC6A2). The first study (
22) found that the T allele of the
SLC6A2 T182C polymorphism was associated with an improved response to milnacipran. The second study (
23) found that variants in the norepinephrine transporter gene (
SLC6A2) predicted response to nortriptyline.
Genome-wide association studies.
Four genome-wide association studies of antidepressant response have been published (
24,
25,
27,
28). This first study involved assessment of individuals from the STAR*D sample who developed treatment-emergent suicidal ideation with citalopram (
24). After the analysis of ∼110,000 single nucleotide polymorphism (SNPs), a single marker mapping to the Papilin (
PAPLN) gene was significant after correction for multiple testing. Little is known about
PAPLN other than that it is a proteoglycan-like sulfated glycoprotein; it is unclear how this gene influences suicidal outcomes in citalopram treatment.
The second study again used the STAR*D cohort to examine the efficacy response to citalopram considering alleles at ∼430,000 variants (
25). Although no variants reached genome-wide significance after correction for multiple testing, three loci showed suggestive evidence of being involved in treatment response. Variants near the ubiquitin protein ligase E3C (
UBE3C), bone morphogenetic protein 7 (
BMP7), and retinoic acid-binding receptor α (
RORA). Although it is unclear how these novel genes might be involved in MDD, it is interesting to note that RORA might be involved in pathways integral to circadian rhythms in mammalian systems (
26).
The third study used cohorts collected in the Munich, Germany, Antidepressant Response Signature (MARS) project that examined drug efficacy after antidepressants were prescribed according to doctor choice. The authors report that they mapped an aggregate series of markers that predict low versus high response to various drug treatments (
27). This finding suggests that treatment response may be multifactorial and under the control of a number of additive genetic loci instead of a limited number with large effects.
The fourth genome-wide association study used samples from the Genome-Based Therapeutic Drugs for Depression (GENDEP) study, which is a partially randomized open-label pharmacogenetic trial (
28). In this study, two intergenic regions containing copy number variants on chromosomes 1 and 10 were associated with the outcome of treatment with escitalopram or nortriptyline at suggestive levels of significance. Drug response to nortriptyline was best predicted by variation in the uronyl 2-sulfotransferase gene. Response to escitalopram was best predicted by a marker in the interleukin-11 (
IL11) gene.