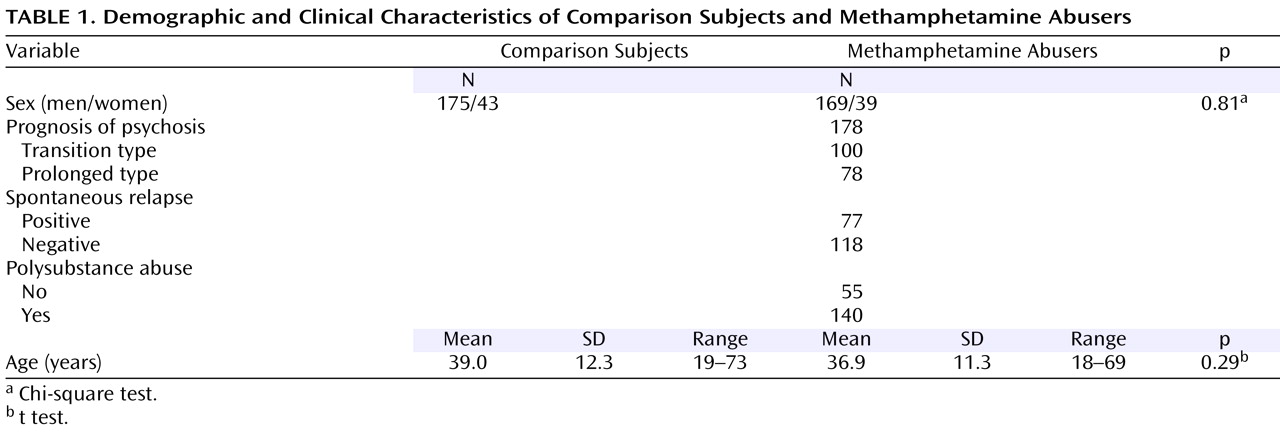
Identification of SNPs and Association Studies
In searching the transcription start position, we found that exon 1 turned out to stretch beyond the position reported in the public database (
Figure 2 ). Namely, we found that the transcription start position was at 113958, which is 513 bp before the start position (114471) reported in AL031587 (http://www.ncbi.nlm.nih.gov/).
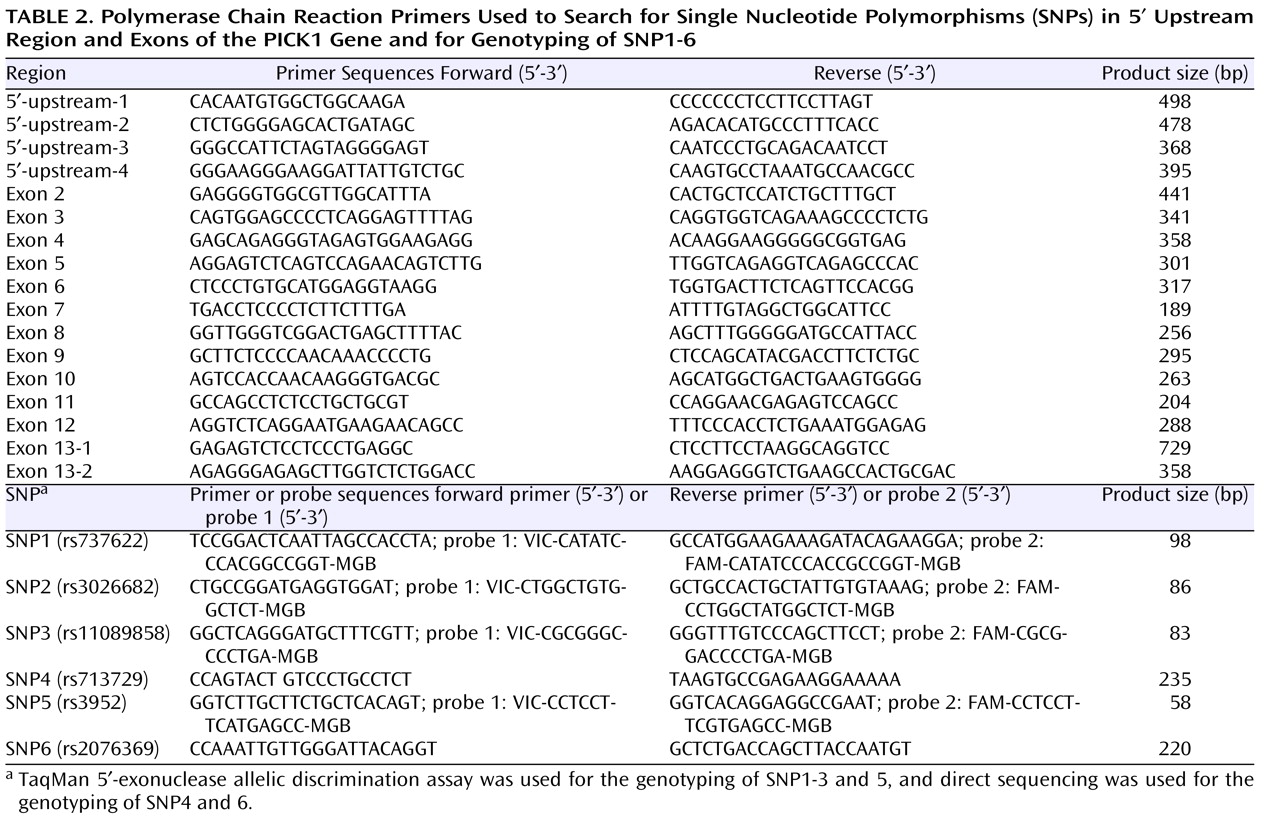
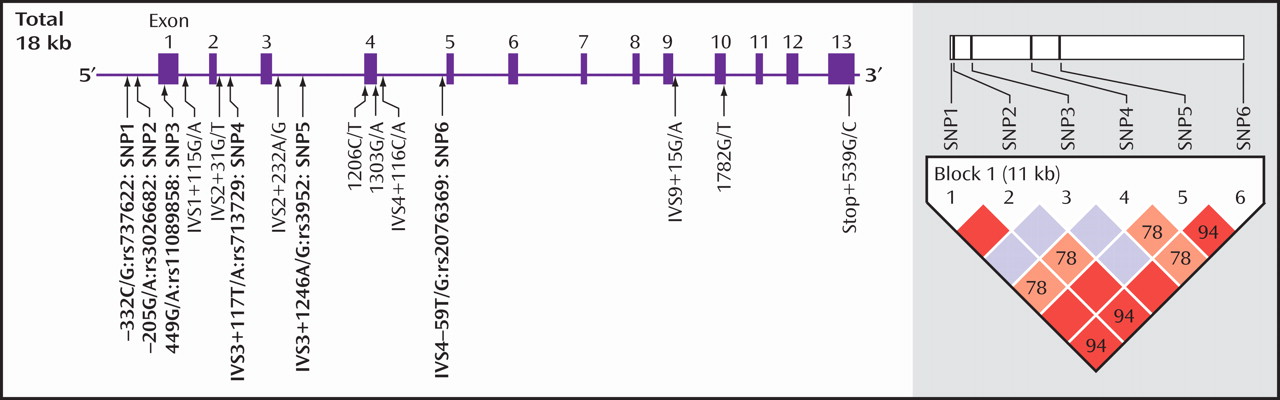
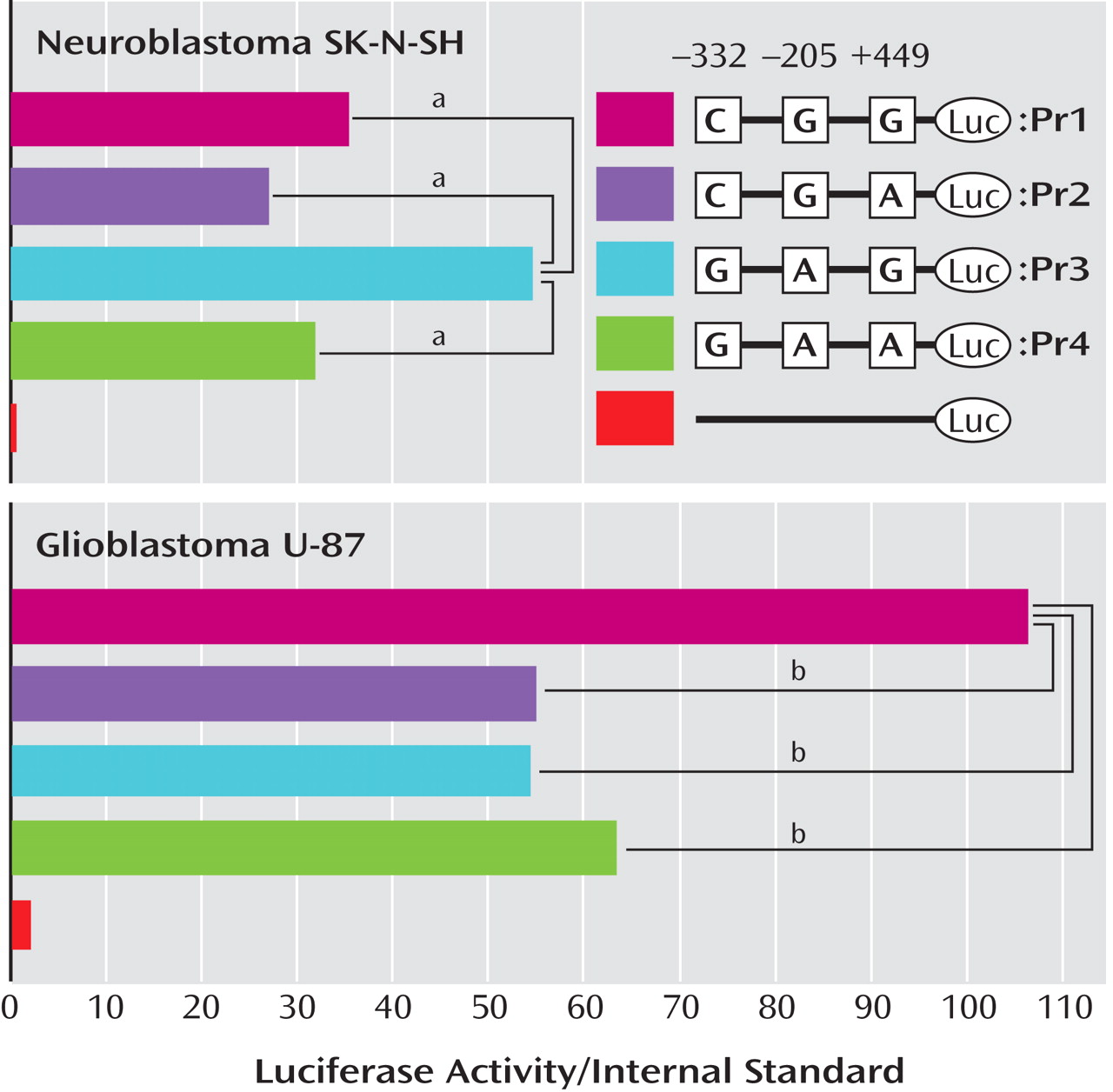
We searched for the SNPs in the PICK1 gene, including the promoter region approximately 500 bp ahead of the transcription start position, the entire 5′-untranslated sequence from the translation start position in exon 2, and all 13 exons and their neighboring sequences. In this study, we found 14 SNPs in the PICK1 gene (
Figure 1 ). Of these SNPs, rs737662 (-332C/G: SNP1), rs3026682 (-205G/A: SNP2), rs11089858 (449 G/A: SNP3), rs713729 (IVS3+117T/A: SNP4), and rs2076369 (IVS4-59T/G: SNP6) were found to be highly frequent (the minor allele >10%) (
Figure 1 ). Subsequent genotyping was performed for these five SNPs (SNP1, 2, 3, 4, and 6) and rs3952 (IVS3+1246A/G: SNP5). Both the genotype and the allele distributions of SNP1, SNP2, and SNP5 were completely the same (
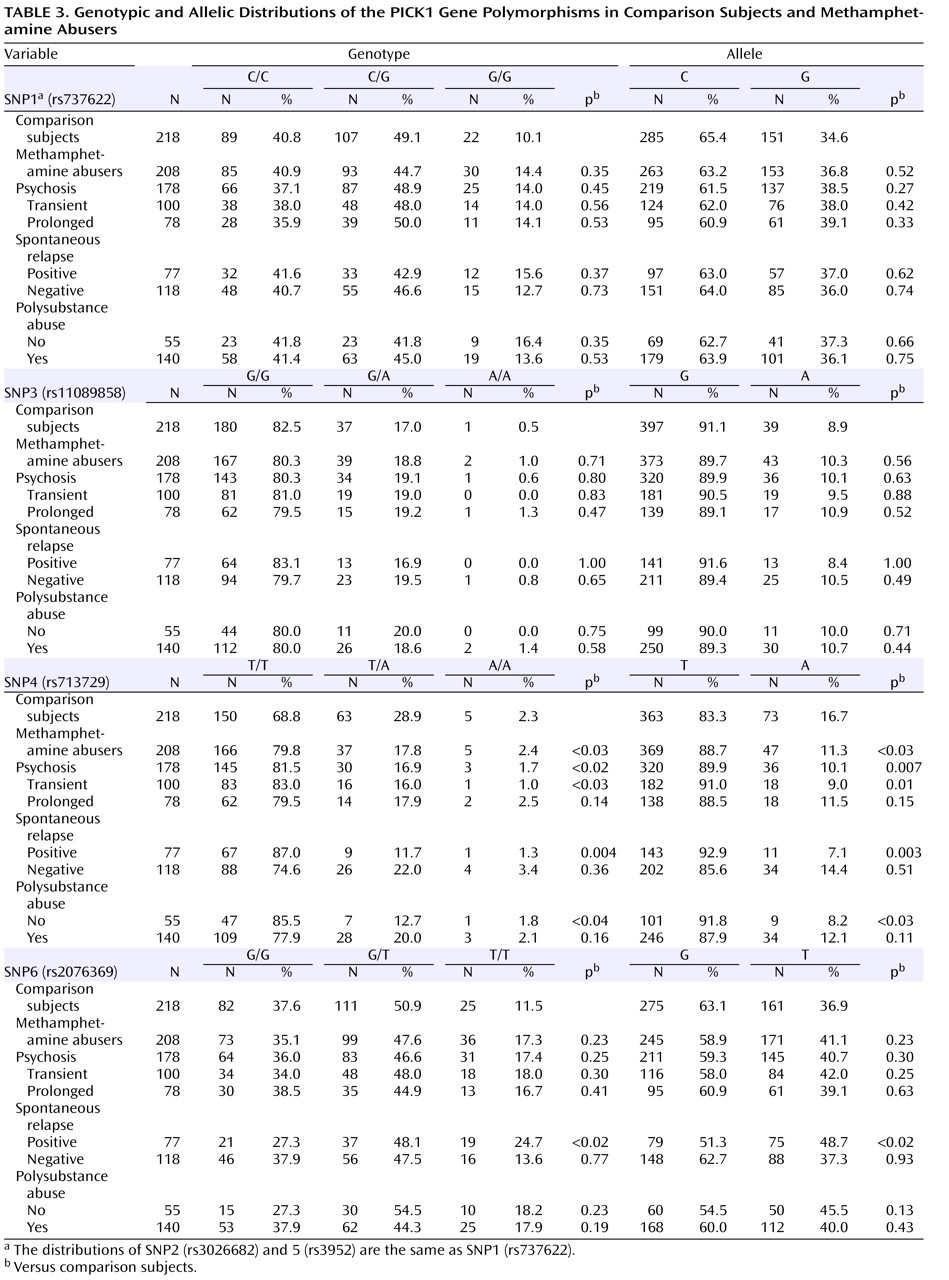
Table 3 ). The allele frequencies and genotype distributions of SNP1, 3, 4, and 6 in methamphetamine abusers and comparison subjects are shown in
Table 3 . The genotype distributions were within the Hardy-Weinberg equilibrium.
We found significantly different frequencies between comparison subjects and methamphetamine abusers in SNP4 (
Table 3 ). The frequency (88.7%) of carrying the T allele among the methamphetamine abusers was significantly higher (odds ratio=1.58, 95% confidence interval [CI]=1.06–2.34, p<0.03) than that of the comparison subjects (83.3%), and we also detected a different distribution of genotype (p<0.03). Positive associations were detected in the subgroup of those who experienced psychosis (alleles, p=0.007, odds ratio=1.79, 95% CI=1.17–2.74, genotype, p<0.02), transient-type psychosis (alleles, p=0.01, odds ratio=2.03, 95% CI=1.17–3.51, genotype, p<0.03), and psychosis with spontaneous relapse (alleles, p=0.003, odds ratio=2.61, 95% CI=1.35–5.07, genotype, p=0.004) and in abusers without polysubstance abuse (alleles, p<0.03, odds ratio=2.26, 95% CI=1.09–4.67, genotype, p<0.04) (
Table 3 ). For SNP6, the frequency (48.7%) of the T allele among methamphetamine abusers who experienced psychosis with spontaneous relapse was significantly higher (odds ratio=1.62, 95% CI=1.19–2.35, p<0.02) than that of the comparison subjects (36.9%), and we also detected a different distribution of genotype (p<0.02) (
Table 3 ). In contrast, no differences for SNP1, 2, 3, and 5 were detected between methamphetamine abusers and comparison subjects (
Table 3 ).
As shown in
Figure 1, a strong linkage disequilibrium was observed in five of these six SNPs. Two haplotypes, C(SNP1)-G(SNP2)-G(SNP3)-A(SNP4)-A(SNP5)-G(SNP6) and G(SNP1)-A(SNP2)-G(SNP3)-T(SNP4)-G(SNP5)-T(SNP6), were significantly different between comparison subjects and methamphetamine abusers (
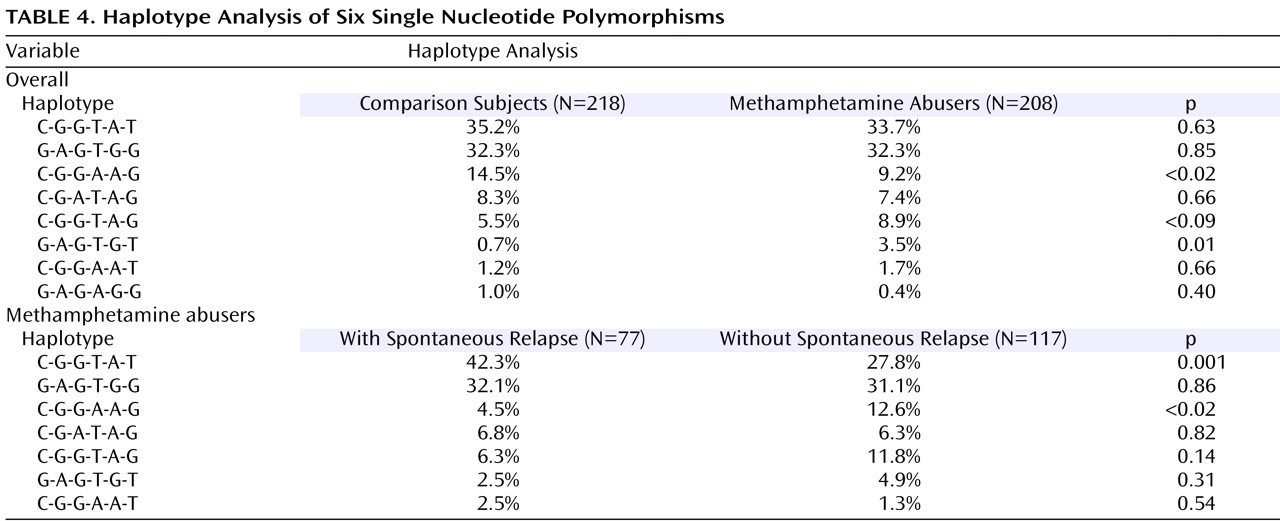
Table 4 ). The frequency (9.2%) of the CGGAAG haplotype in the methamphetamine abusers was significantly lower (odds ratio=0.60, 95% CI=0.45–0.79, p<0.02) than that of the comparison subjects (14.5%), and the frequency (3.5%) of the GAGTGT haplotype in the methamphetamine abusers was significantly higher (odds ratio=5.2, 95% CI=2.27–11.6, p=0.01) than that (0.7%) of the comparison subjects (
Table 4 ). Of interest, a haplotype analysis between methamphetamine abusers with and without spontaneous relapse of psychosis showed the significant difference in the most major haplotype (CGGTAT) as well as the CGGAAG type. The frequency (42.3%) of CGGTAT type in the methamphetamine abusers with spontaneous relapse was significantly higher (odds ratio=2.2, 95% CI=1.80–2.61, p=0.001) than that in those without spontaneous relapse (27.8%) (
Table 4 ). As to the frequency of the CGGAGG type, the frequency (4.5%) in methamphetamine abusers with spontaneous relapse was significantly lower (odds ratio=0.33, 95% CI=0.23–0.47, p<0.02) than that in those without spontaneous relapse (
Table 4 ).