Schizophrenia is a severe psychiatric disorder with a lifetime prevalence of approximately 1% (
1). Like other common, complex disorders, schizophrenia is a multifactorial polygenic disorder that reflects the combined influence of both genetic and nongenetic factors (
1,
2). Epidemiological studies indicate a heritability of up to 80% for schizophrenia, reflecting a strong genetic influence (
3). Several candidate genes have been proposed but as yet remain unconfirmed as harboring causal mutations (
4,
5). Linkage studies of schizophrenia have implicated many chromosomal regions (
6), yet the identification of causative genes within the linked regions has proven difficult. This difficulty in translating linkage findings to causal genes may be due to several factors, including the modest nature of the linkage signals and the broad genetic regions they encompass, genetic heterogeneity, polygenic inheritance, and environmental influences associated with schizophrenia. Additionally, schizophrenia is a highly heterogeneous disorder, with patients exhibiting a broad range of deficits and symptom severity subsumed under a single categorical diagnosis, labeled by Bleuler as the “group of schizophrenias” (
7).
Investigations of schizophrenia and other common disorders have recently moved toward genome-wide association studies (GWAS), which have greater power to detect weak associations to common variants. However, a considerable proportion of the observed heritability is not detectable in these large studies of unrelated patients and comparison subjects (
11,
12). One likely source of this “missing heritability” is allelic heterogeneity, which may substantially reduce the power of GWAS to detect causative genes because the overall genetic effect is divided among multiple variants, some of which may be rare and not well captured by common tag single-nucleotide polymorphisms (SNPs). Linkage, however, can detect the aggregate effects of multiple rare and common variants within a susceptibility gene or region, even with different mutations conferring risk in different families. Linkage data may also be used to weight evidence for association and potentially increase the power of GWAS (
13). Thus, linkage studies and family-based samples continue to be relevant in the age of rapidly advancing technologies.
The Consortium on the Genetics of Schizophrenia (COGS) focuses on investigating endophenotypes as a strategy for dissecting the genetic architecture of schizophrenia and filling in the “gene to phene” gap (
8–
10). Twelve heritable neurophysiological and neurocognitive endophenotypes that are characteristically impaired in schizophrenia patients were chosen for the COGS study: prepulse inhibition of the startle response (
14–
16), P50 event-related potential suppression (
17–
19), the antisaccade task for eye movements (
20,
21), the Continuous Performance Test (degraded-stimulus version) (
22,
23), the California Verbal Learning Test, 2nd edition (
24,
25), the Letter-Number Span (
26–
28), and six measures from the University of Pennsylvania Computerized Neurocognitive Battery (abstraction and mental flexibility [
29,
30], face memory [
29,
30], spatial memory [
29,
30], spatial processing [
29,
30], sensorimotor dexterity [
29,
30], and emotion recognition [
29,
30]). Deficits in all endophenotypes have been demonstrated not only in patients with schizophrenia but also in their clinically unaffected relatives, suggesting that these deficits reflect part of the heritable risk for the illness. Complete reviews of each endophenotype in the COGS study, including the rationale for selection and data regarding stability, reliability, and heritability, have been reported elsewhere (
31–
33). We previously reported evidence of significant heritability for these 12 endophenotypes in a subset of 183 COGS families and have demonstrated association with several carefully chosen candidate genes in 130 of these families (
34,
35). In the present study, we report the results of a genome-wide SNP linkage scan for these endophenotypes in the complete COGS sample of 296 families.
Results
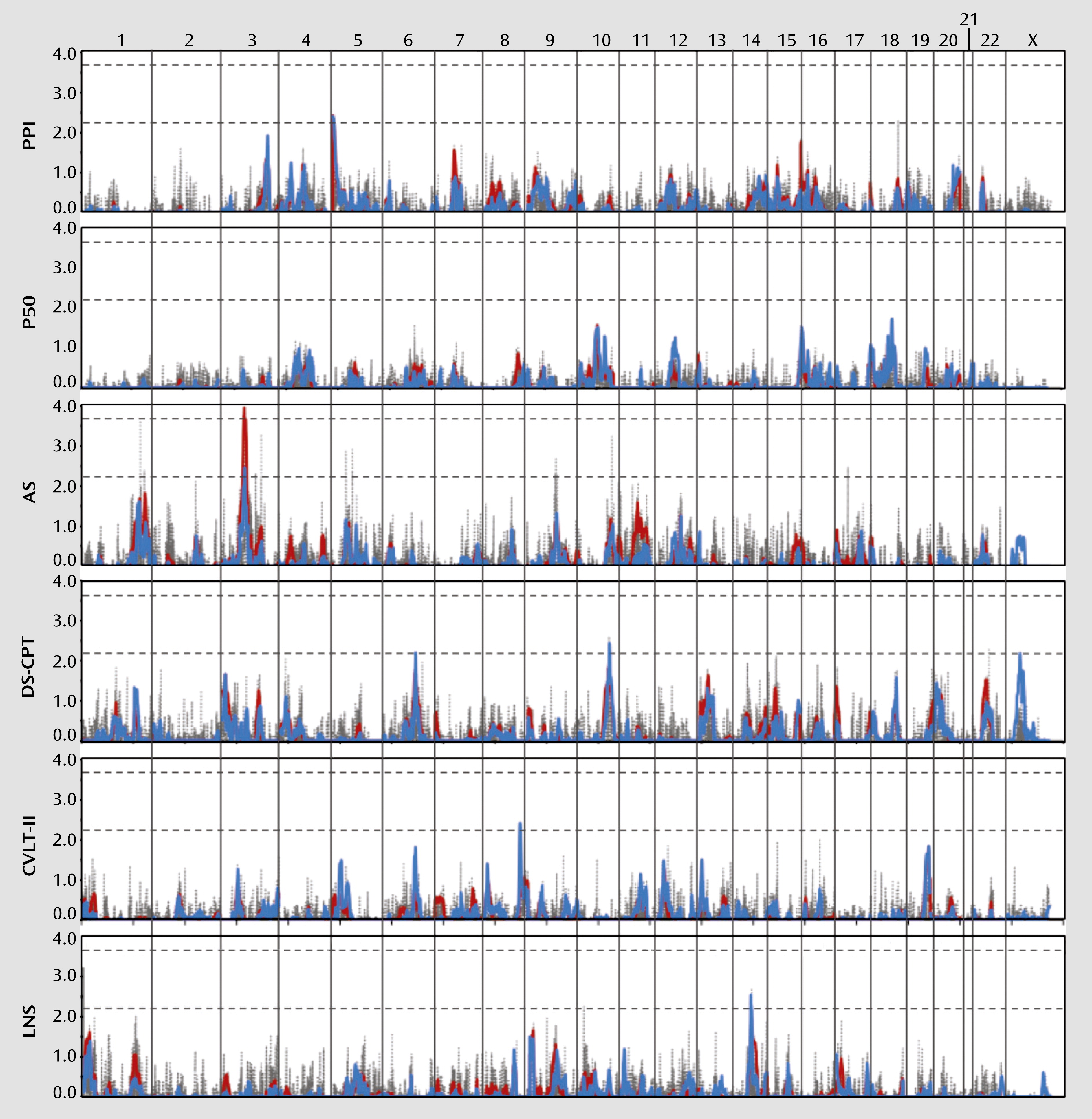
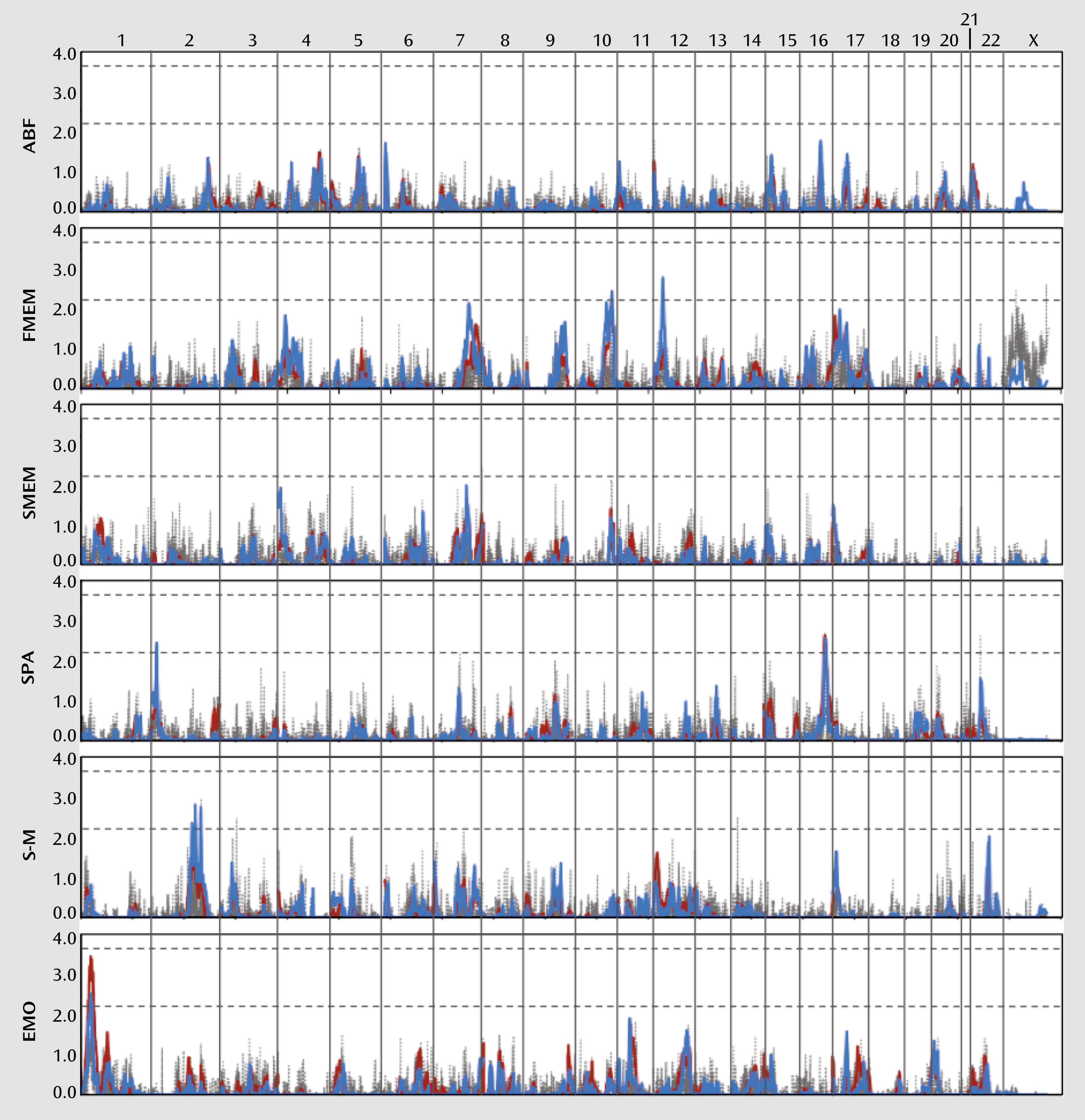
We performed a genome-wide linkage scan for each endophenotype using variance components methods as the primary analysis. Regression-based methods were then used to confirm and extend the results. As shown in
Figures 1 and
2 and summarized in
Table 3, these analyses have collectively identified several linkage regions meeting at least suggestive evidence of linkage across the 12 endophenotypes, according to the criteria established by Lander and Kruglyak (
50). A summary of all multipoint LOD scores >1.0 with the corresponding empirical p values is provided in Table S2 in the online data supplement, and a complete listing of all multipoint LOD scores is available in Table S3 in the data supplement.
Significant evidence for linkage (LOD score >3.6) was observed for the antisaccade task on chromosome 3p14, with a variance components LOD score of 4.0. While the regression LOD score for this region only reached suggestive evidence for linkage, with a peak LOD score of 2.4, simulation analyses indicated an empirical p value <0.0001. No other endophenotype displayed linkage to this region with a LOD score >1.0 (see Table S2 in the online data supplement). Several neuronally expressed genes are located beneath this linkage peak, including ataxin 7 (
ATXN7), which encodes a protein involved in chromatin remodeling and plays a role in transcriptional regulation. A polyglutamine repeat expansion in this gene is implicated in spinocerebellar ataxia type 7, which also presents with retinal degeneration and visual loss, dementia, hypoacusia, severe hypotonia, and auditory hallucinations (
51). The
ATXN1 gene on chromosome 6p22, which causes spinocerebellar ataxia type 1 through a similar mechanism, has also been investigated as a candidate gene with associations to schizophrenia (
52,
53).
Another region nearly reaching genome-wide significance under the variance components model was chromosome 1p36, which produced a LOD score of 3.5 for emotion recognition. The regression LOD score for this region only reached 2.5, yet simulation analyses indicated an empirical p value <0.0001. The Letter-Number Span also revealed modest evidence for linkage to this region, with a LOD score of 1.6 (see Table S2 in the online data supplement). Several genes are located beneath this peak, including the serotonin receptor 6 gene (
HTR6), which functions in the modulation of cholinergic and dopaminergic neurotransmission, plays a role in spatial learning and memory, and has a high affinity for several conventional and atypical antipsychotics (
54).
Suggestive evidence for linkage (LOD score >2.2) under the variance components model was observed for prepulse inhibition on chromosome 5p15, with a peak LOD score of 2.5 (regression LOD score=2.4); for face memory on chromosome 10q26, with a peak LOD score of 2.3 (regression LOD score=2.4); and for spatial processing on chromosome 16q23, with a peak LOD score of 2.6 (regression LOD score=2.5). The regression method identified several additional regions meeting suggestive evidence for linkage, most of which displayed at least some evidence for linkage, with LOD scores >1.0 in the variance components analysis. These regions included 2p25 (spatial processing), 2q24 and 2q32 (sensorimotor dexterity), 8q24 (California Verbal Learning Test), 10q26 (degraded-stimulus Continuous Performance Test), 12p12 (face memory), and 14q23 (Letter-Number Span).
Several genes of potential interest were identified beneath the suggestive peaks. The gene encoding the zinc finger protein 804A (
ZNF804A) lies beneath the linkage peak for sensorimotor dexterity on chromosome 2q32. This gene has shown strong evidence for association with schizophrenia in several large GWAS (
55–
57). Although the region on chromosome 5p15 with linkage to prepulse inhibition is very gene-dense, one gene of particular note, the dopamine transporter gene (
SLC6A3/
DAT), lies beneath the peak. This gene has shown evidence of association with prepulse inhibition and startle habituation (
58), schizophrenia (
59), and several of our neurocognitive endophenotypes (
35). Prepulse-inhibition deficits have also been reported in
DAT knockout mice (
60). Finally, the glutamate (N-methyl-
d-aspartic acid) receptor 2B gene (
GRIN2B) lies beneath the linkage peak for face memory on chromosome 12p12. Several studies have found evidence for association of
GRIN2B with schizophrenia (
58,
61–
63), and we previously reported associations with several of our neurocognitive endophenotypes (
35,
58).
We also identified many regions of coincident linkage in which at least two endophenotypes produced modest evidence for linkage (LOD score >1.0), including the 1p36, 10q26, and 12p12–13 regions described above (see Table S2 in the online data supplement). Two regions in particular on 10q26 and 17p13 revealed linkage to multiple endophenotypes, some of which were genetically correlated (see Table S1 in the data supplement). A multivariate linkage analysis combining the antisaccade task, degraded-stimulus Continuous Performance Test, face memory, and spatial memory produced a LOD score of 2.1 for 10q26. However, face memory alone produced suggestive LOD scores of 2.2 and 2.4 with the two linkage methods, and the degraded-stimulus Continuous Performance Test also produced a suggestive LOD score of 2.4 with the regression method. It is thus likely that these linkages represent distinct signals despite their close proximity. A similar analysis was conducted for chromosome 17, combining the degraded-stimulus Continuous Performance Test, face memory, spatial memory, and sensorimotor dexterity. While individual endophenotypes produced LOD scores of 1.3–1.8 for 17p13 under the variance components model, a suggestive LOD score of 2.2 was observed through their joint analysis, which may indicate the presence of a gene in this region that is involved in some aspect of neurocognition or one that generally contributes to schizophrenia susceptibility. Notably, the
YWHAE gene lies below this peak and encodes 14–3-3epsilon, a member of a highly conserved family of proteins involved in a wide range of signaling pathways.
YWHAE is a binding partner of
DISC1 and has been proposed as a susceptibility gene for schizophrenia (
64,
65).
Discussion
Investigations of endophenotypes that reflect aspects of the brain pathology involved in schizophrenia may facilitate the identification of genes contributing to schizophrenia susceptibility (
8–
10). Genome-wide linkage analyses of 12 heritable neurocognitive and neurophysiological endophenotypes collectively identified 12 regions displaying genome-wide significant or suggestive evidence for linkage using two complementary linkage analysis methods. Several genes of potential interest are located beneath these linkage peaks, including
HTR6 on chromosome 1p36 (emotion recognition),
ZNF804A on 2q32 (sensorimotor dexterity),
ATXN7 on 3p14 (the antisaccade task),
DAT on 5p15 (prepulse inhibition),
GRIN2B on 12p12 (face memory), and
YWHAE on 17p13 (multivariate cognitive phenotype).
We did not find evidence for linkage of the COGS endophenotypes to some of the prominent linkage regions for schizophrenia, such as 1q21–22, 4q31, 5q22–31, 6p22–24, 8p21–22, 9q21–22, and 10p11–15. However, we did find at least modest evidence to support linkage with LOD scores >1 observed for at least one endophenotype to other linkage regions identified for schizophrenia (
6), such as 1q32–41, 5p13–14, 6q21–22, 13q14–32, 15q13–15, 22q11–13, and Xp11 (see Table S3 in the online data supplement). A SNP linkage study of schizophrenia, conducted by Holmans et al. (
66), identified additional suggestive linkage regions on chromosomes 8q24, 9q34, and 12q24, while another study found suggestive evidence for linkage of schizophrenia covaried for positive symptom dimensions on chromosomes 2q32, 10q26, and 20q12 (
67). In our study, we also observed suggestive evidence for linkage to 2q32 (sensorimotor dexterity), 8q24 (the California Verbal Learning Test), and 10q26 (the degraded-stimulus Continuous Performance Test and face memory).
Few studies investigating schizophrenia endophenotypes through genome-wide analyses have been published to date. One study of several measures of neurocognition in schizophrenia identified a linkage peak that reached genome-wide significance on chromosome 12q24 for undegraded Continuous Performance Test hit rate (
68). We identified modest evidence for linkage to this region for one of our neurocognitive endophenotypes (emotion recognition). In another study of multigenerational schizophrenia families with theoretically greater genetic loading for both schizophrenia and associated endophenotype deficits, significant evidence of linkage was observed for schizophrenia on 19q13 and abstraction and mental flexibility on 5q15 (
69). Modest evidence for linkage was observed in our sample for neurocognitive endophenotypes in these regions, including abstraction and mental flexibility on 5q15.
We also identified several regions of overlapping linkage signals of two or more endophenotypes, some of which have featured prominently in previous linkage studies of schizophrenia, such as 6q21–22, 15q13–15, and 22q11–13 (
70–
77). The 15q13–15 region was identified as a susceptibility locus for schizophrenia through a linkage study of P50 suppression and contains the alpha-7 nicotinic acetylcholine receptor gene (
CHRNA7), a candidate gene for schizophrenia (
18). Deletions within the 22q11 region have been associated with schizophrenia, and two prominent candidate genes, catechol-
O-methyltransferase (
COMT) and proline dehydrogenase (
PRODH), are located in this region (
78). A meta-analysis combining the results of several linkage studies confirmed this region as a valid linkage region for both schizophrenia and bipolar disorder that likely contains one or more susceptibility genes (
79), and evidence of linkage for a composite inhibitory phenotype combining P50 suppression and antisaccade was observed for 22q11–12 (
80). Finally, recent studies suggest that rare deletions in the 15q13 and 22q11 regions predispose to schizophrenia (
81–
84). Thus, these regions may warrant further investigation in other samples.
Recent GWAS have identified several risk genes associated with schizophrenia at genome-wide significance levels in very large samples. These include microRNA 137 (MIR137) on chromosome 1p21,
ZNF804A on 2q32, the major histocompatibility complex region on 6p21–22, CUB and sushi multiple domains 1 (
CSMD1) on 8p23, the neurogranin gene (
NRGN) on 11q24, and transcription factor 4 (
TCF4) on 18q21 (
55–
57,
85,
86). The voltage-dependent
l-type calcium channel alpha-1C (
CACNA1C) on chromosome 12p13 also reached genome-wide significance in a joint analysis with bipolar disorder (
85). Other than the aforementioned suggestive evidence of linkage for sensorimotor dexterity to the 2q32 region containing
ZNF804A, we observed only modest evidence of linkage for the 8p23 (the California Verbal Learning Test), 12p13–12 (abstraction and mental flexibility, sensorimotor dexterity), and 18q21 (P50 suppression) regions.
One might expect that the endophenotypes with the highest heritabilities would produce the strongest genetic signals, yet, as our results demonstrate, this is not always the case. In our study, antisaccade had one of the highest heritabilities, at 36%, and produced the only genome-wide significant linkage signal, whereas spatial memory did not produce a linkage signal meeting genome-wide suggestive criteria, despite having a comparable heritability of 34%. The second strongest linkage signal was observed for emotion recognition, the endophenotype with the lowest heritability in this study at 16%, although we do note that emotion recognition demonstrated a much higher heritability of 32% in our previous analysis of 183 families (
34). Although it is possible that the genetic signal from the original families is strong enough to influence the overall linkage signal for emotion recognition, these results may simply illustrate that heritability estimates are not perfect predictors of the potential “mapability” of the underlying genetic variants. An endophenotype with a relatively low but significant heritability may exhibit large effects of a small number of genes, which would facilitate mapping. Alternatively, an endophenotype may be highly heritable but also highly polygenic, similar to schizophrenia itself, which would significantly complicate gene mapping by producing low-level signals across a multitude of genomic regions. For example, a recent study evaluating the variance in liability explained by the identified variants for 10 complex diseases, including schizophrenia, found that each associated SNP explained a median variance of only 0.25% (
87). While endophenotypes provide reliable measures of specific neurophysiological and neurocognitive processes that are deficient in schizophrenia, they may not exhibit simpler genetic architectures (
88).
There are several limitations or caveats applicable to this study. First, the COGS family ascertainment strategy focused on the recruitment of siblings discordant for schizophrenia to increase variation in the endophenotypic values. As a result, only 16 affected sibling pairs and eight affected parents exist in the sample, too few to reliably assess linkage to schizophrenia or determine whether the genomic regions identified by the endophenotypes have an effect on schizophrenia susceptibility. We were also unable to assess the degree of genetic correlation between the endophenotypes and schizophrenia. Additionally, this ascertainment scheme may result in the underestimation of endophenotype heritabilities and genetic correlations to the extent that they are correlated with schizophrenia. Second, with a linkage study of 12 endophenotypes, multiple comparisons are an issue. It is difficult to determine the appropriate correction in this case, since most of the endophenotypes are significantly correlated with each other (
34). Finally, our sample of predominantly small nuclear families lacks sufficient power to reliably detect loci with smaller effects in a linkage analysis. The relatively low power of this study is independent of the heritabilities of these endophenotypes and in part reflects variance in the available data across endophenotypes. Yet, we successfully identified several genomic regions that warrant further investigation, despite being underpowered.
We embarked on this endophenotype strategy to provide a platform to dissect the polygenic basis of schizophrenia susceptibility and to identify therapeutic molecular targets for the treatment of schizophrenia. In this context, endophenotypes were used as a complementary strategy to augment the dissection of the clinical and genetic heterogeneity of schizophrenia, as we and other investigators have discussed (
8–
10). The linkage analyses of the 12 endophenotypes described in this study represent a first step toward this goal. While the absence of significant linkage signals for all endophenotypes may be an issue of power deficiencies, it is also likely a reflection of the genetic complexity of the endophenotypes. For example, abnormalities in at least 20 different brain regions have been identified in various schizophrenia cohorts, five of which fall within regions known to regulate mammalian prepulse inhibition (
89,
90). Deficient prepulse inhibition in schizophrenia might arise from any of these neural abnormalities, which almost certainly reflect a heterogeneous group of genetic determinants. Thus, while the biological basis of these endophenotypes may be simpler than that of schizophrenia per se, they nonetheless remain complex and appear to be highly polygenic. Conceivably, a refinement of these endophenotypes may probe a more specific physiology and thereby be sensitive to a more pure genetic signal. Despite these complexities, we have identified several regions meeting the standard genome-wide significant and suggestive criteria that provide further support for several existing schizophrenia candidate genes and chromosomal regions. The extent to which these regions harbor specific mutations that are involved in schizophrenia susceptibility, or the cognitive and neurophysiological processes tapped by our endophenotypes, remains a topic for future discussion to be informed by ongoing assessments of copy number variation burden and methylation events, as well as future sequencing efforts. While schizophrenia and its treatment will not be easily resolved, the use of interlocking genomic and endophenotype approaches offers much hope for the future (
91).