Late-life depression is a disabling mental disorder affecting more than 2 million older adults in the United States (
1). More than 50% of patients with late-life depression do not achieve remission with an antidepressant trial (
1). In older adults, prolonged depressive symptoms are associated with greater caregiver burden, medical complications, functional impairment, and cognitive decline (
2). Therefore, finding predictive biomarkers for antidepressant treatment success in older adults is imperative to achieve timely remission (
3). Only a few genetic polymorphisms have been consistently investigated for associations with antidepressant response, resulting in varying levels of evidence for genetic biomarkers (
4); therefore, more research is needed in this area.
Venlafaxine is a serotonin-norepinephrine reuptake inhibitor (SNRI) with evidence supporting its use for the treatment of late-life depression (
5). Venlafaxine is typically prescribed at dosages between 75 and 300 mg/day, which have been correlated with differential binding affinity and inhibitory properties at norepinephrine and serotonin transporters in vivo. Previous investigations have shown that venlafaxine exerts dose-dependent norepinephrine reuptake inhibition; a dosage of 150 mg/day or higher is sufficient to produce noradrenergic activity (
6). Genetic variants across serotonergic and noradrenergic neurotransmitter systems have been investigated for association with venlafaxine treatment response in younger adults, including the serotonin transporter gene (
SLC6A4; 5-HTTLPR [
7–
9], VNTR [
9–
11], and rs25531 [
10,
12]), the serotonin receptor 2A gene (
HTR2A; rs7997012 [
12] and G-1438A [
9,
11]), the tryptophan hydroxylase gene (
TPH; A218C [
9,
13]), and the norepinephrine transporter gene (
NET, SLC6A2; rs5569). Despite these observations in younger adults, to our knowledge, this is the first study to investigate the associations between late-life depression treatment remission variability, venlafaxine dosage, and variants in
NET/SERT genes, as well as other potentially involved serotonergic system genes.
Results
Sample Characteristics
Our final sample included 350 participants of European (N=311, 88.9%), African (N=33, 9.4%), Asian (N=5, 1.4%), and admixed (N=1, 0.3%) ancestries (
Table 1) (see also Figure S1 in the
online data supplement). The sample consisted predominantly of women (N=223, 63.7%), and the mean age was 68.6 years (SD=7.0). At baseline, participants were typically moderately depressed (mean MADRS score, 26.6 [SD=5.6]), and most had recurrent depression (N=255, 72.9%). On average, participants spent 94.6 days (SD=18.4) in the treatment protocol. The mean venlafaxine dosage was 241.6 mg/day (SD=70.3). Overall, 179 participants (51.1%) remitted, and the mean percentage change in MADRS score from baseline to end of treatment was −50.3 (SD=37.4%).
Exploration of Potential Clinical Covariates
Several clinical variables were found to be significantly and independently associated with remission status in the total (mixed ancestry) sample and the European sample. Compared with patients who did not remit, those who remitted were more likely to be older (mean=69.9 years [SD=7.6] compared with mean=67.4 years [SD=6.1]; p=0.001), to be female (χ2=9.63, df=1, p=0.002), to have a shorter duration of illness (mean=227.7 weeks [SD=540.5] compared with mean=430.4 weeks [SD=782.1]; t=2.47, df=335, p=0.014), and to have lower baseline MADRS scores (mean=24.8 [SD=5.3] compared with mean=28.4 [SD=5.4]; t=6.32, df=348, p<0.001). Patients who remitted spent a longer time in the treatment protocol (mean=97.0 days [SD=18.8] compared with mean=92.4 days [SD=18.4]; t=−2.58, df=348, p=0.010). Remission rates also varied across recruitment sites (χ2=8.23, df=2, p=0.016). All variables that were significantly associated with remission were also significantly associated with percentage change in MADRS score and time to remission. Duration of illness (for the current major depressive episode) was dropped as a potential covariate because of a significant correlation with baseline MADRS score (rs=0.13, p=0.017) and a small effect on remission status (β=−0.13, p=0.13). There was no association of ethnicity with remission status, percentage change in MADRS score, or time to remission. Based on the above univariate analysis and removal of significantly correlated covariates, we included in the ANCOVA model age, sex, baseline MADRS score, treatment duration, and site. See Table S3 in the online data supplement for a full description of these covariates across phenotypes.
Norepinephrine and Serotonin Transporter Variants and Remission Status
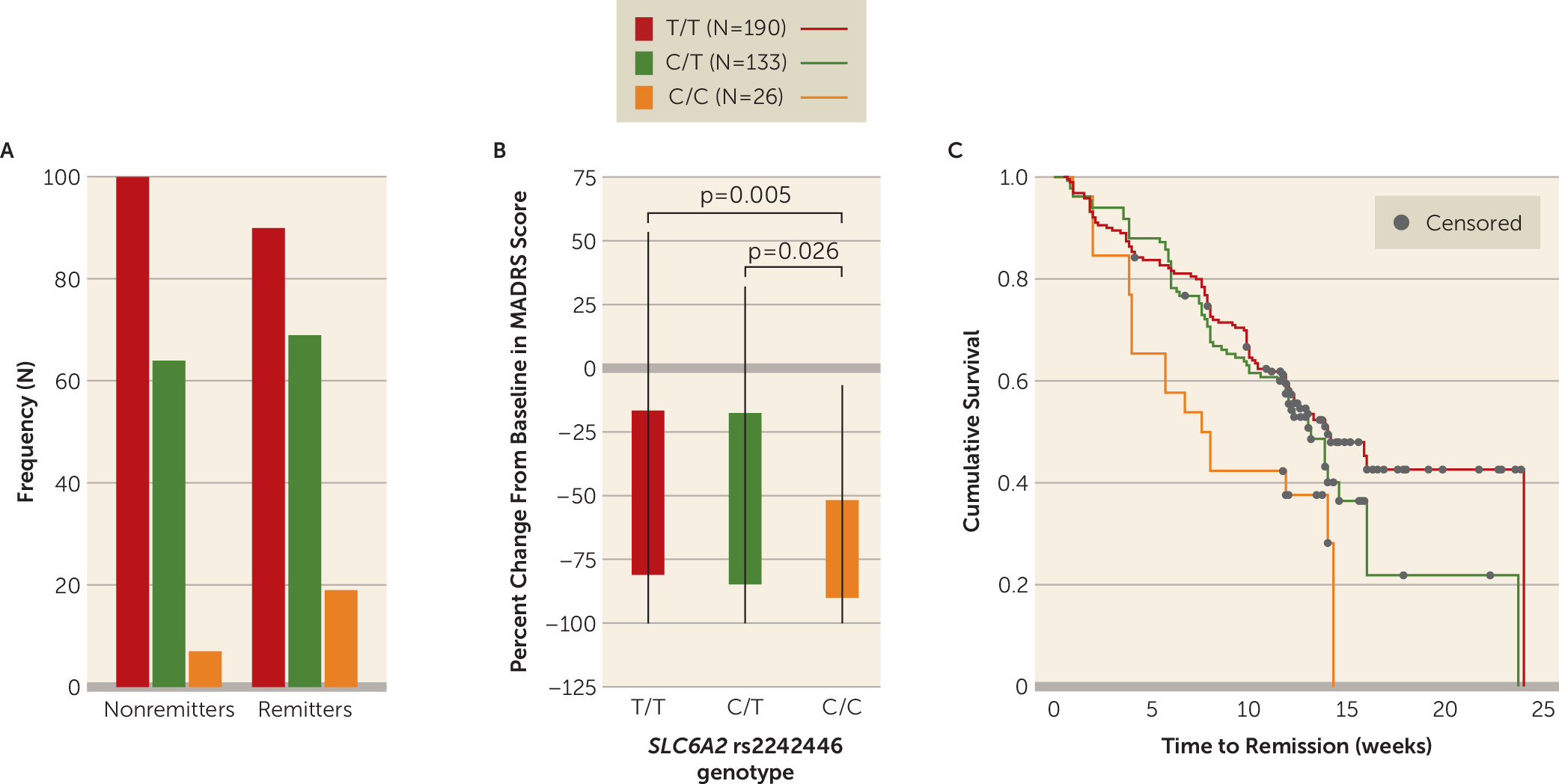
NET (
SLC6A2) variant rs2242446 was significantly associated with remission/nonremission in the total (mixed ancestry) sample (odds ratio=1.66, 95% CI=1.13, 2.42, p=0.009) (
Figure 1) and nominally associated in the European-ancestry subsample (odds ratio=1.67, 95% CI=1.11, 2.46, p=0.013; see Figure S2 in the
data supplement) after adjusting for covariates and multiple comparisons (see Table S4 in the
data supplement for a full breakdown of the logistic regression). In the total sample, individuals with the rs2242446 C/C genotype were more likely to remit (73.1%) than those with either the C/T (51.8%) or the T/T genotype (47.3%). There were no significant differences among rs2242446 genotypes in terms of age, age at onset, duration of illness, duration of treatment, and baseline MADRS score. There was also no significant effect of ancestry (see Table S5 in the
data supplement). The same pattern of association of rs2242446 with remission but lack of genotype differences across other baseline variables were also seen in the European-ancestry subsample but not in the smaller African-ancestry subsample (N=31) (see Table S6 in the
data supplement).
NET variant rs2242446 was also associated with percentage change in MADRS score (partial eta squared [η
2]=0.03, p=0.006) in the total (mixed ancestry) sample and nominally associated in the European-ancestry subsample (η
2=0.028, p=0.013). Furthermore, rs2242446 genotype was associated with time to remission in the total sample (Mantel-Cox χ
2=9.47, p=0.009) and nominally associated in the European-ancestry sample (χ
2=7.84, p=0.02). In the total sample, individuals with the C/C genotype had a shorter time to remission (mean=8.13 weeks [SD=4.64]) than those with the C/T (mean=10.20 weeks [SD=4.19], p=0.089) or T/T (mean=10.65 weeks [SD=4.84], p=0.023) genotypes. In linear mixed-model analyses,
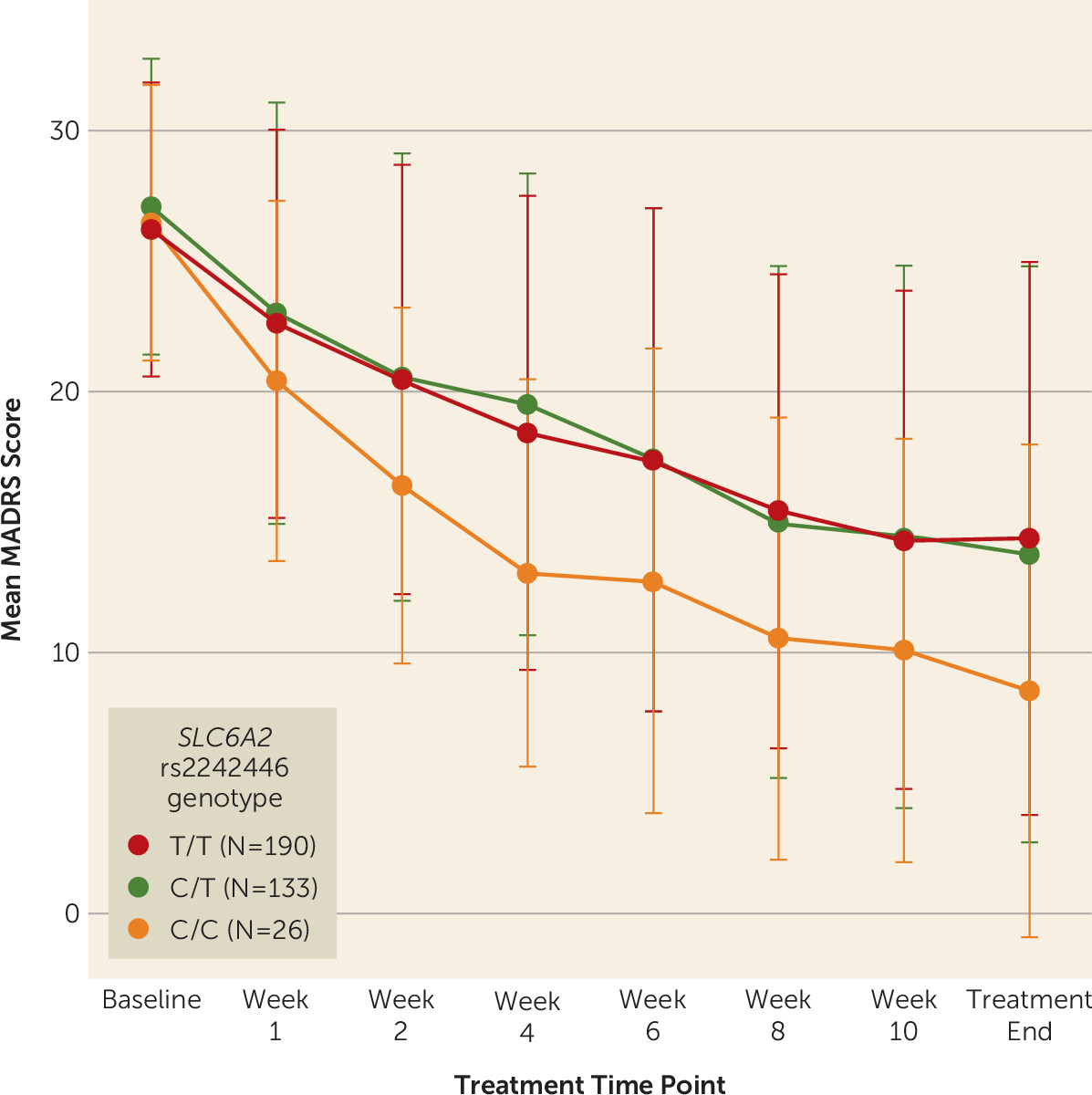
NET variant rs2242446 was also nominally associated with change in MADRS score across treatment time points (F=8.08, df=2, p=0.018) before correction for multiple testing. After only 2 weeks of treatment, individuals with the rs2242446 C/C genotype already showed a nominally significant greater reduction in MADRS score (mean change, −38.84% [SD=22.42) than those with the C/T (mean change, −24.51% [SD=26.66]; p=0.034) or T/T genotypes (mean change, −22.46% [SD=27.20]; p=0.010) (
Figure 2). Given that individuals with the C/C genotype showed earlier and better response, they also required a significantly lower dosage of venlafaxine on average (mean=209.1 mg [SD=78.8]) at the end of treatment than those with the T/T genotype (mean=248.1 mg [SD=67.4]; p=0.022) but not those with the C/T genotype (mean=239.4 mg [SD=71.2], p=0.11).
For the serotonin transporter, we observed no significant associations of rs25531/5-HTTLPR and 5-HTTVNTR genotypes with our outcomes of interest in the total (mixed ancestry) sample and the ethnic (European and African) subsamples (p>0.05). Similarly, there were no significant associations with time to remission or across treatment time points.
Other Serotonergic System Variants
Other investigated serotonergic system genes showed inconsistent association across several outcomes before correction for multiple testing.
HTR2A variant rs6311 was nominally associated with percentage change in MADRS score in the total sample (η
2=0.018, p=0.047) and the European-ancestry (η
2=0.020, p=0.044) sample. However, rs6311 was not associated with response across time points, remission, or time to remission in any sample. Similarly, none of the other serotonergic system genes (
HTR1A,
HTR1B,
HTR2C,
TPH1, and
TPH2) showed any consistent significant associations across multiple outcomes (
Table 2) (see also Table S6 in the
online data supplement).
Functional Effects of rs2242446 Neural NET Expression
In order to understand the functional effects of rs2242446 on the expression of the
NET gene across different brain regions, we conducted gene expression analyses (
35) using data from the UK Brain Expression Consortium (BRAINEAC;
http://braineac.org). Using BRAINEAC data, we found that rs2242446 genotypes differentially affect
NET gene expression within the hippocampus (p=0.037), the thalamus (p=0.037), and the temporal cortex (p=0.048) (see Figure S3A in the
data supplement). In these regions, individuals with the C/C genotype show significantly lower expression of
NET than do either those with the C/T genotype or those with the T/T genotype.
Discussion
This study is, to our knowledge, the first to examine the association between NET and serotonergic system gene polymorphisms in association with venlafaxine remission in late-life depression, using a large and well-characterized sample. We found that the NET gene variant rs2242446 (T-182C) was associated with multiple measures of treatment success in older depressed adults treated with venlafaxine: individuals with the C/C genotype had greater odds of remission, shorter time to remission, and a greater percentage change in depressive symptoms than those with either the T/T or the C/T genotype.
Strikingly, the association of NET rs2242446 with remission was also accompanied by an earlier response. Given that the majority of individuals with the C/C genotype received at least 150 mg/day at the end of treatment (92.3%, N=24), there is reasonable evidence suggesting that the norepinephrine transporter was being sufficiently inhibited to produce clinically meaningful effects. Because of their favorable response, individuals with the C/C genotype required lower dosages of venlafaxine on average. Since response to venlafaxine is likely influenced by additional factors, our findings warrant further investigation of NET to assess the clinical utility of pharmacogenetic testing in older adults. In particular, this preliminary finding requires replication in comparable independent samples to determine its potential to be integrated into currently available genetic algorithms.
There is functional evidence to suggest that
NET variant rs2242446 may play a significant role in the regulation of
NET expression. Located 1.1 kb upstream of the
NET transcription start site, rs2242446 has promoter histone markers and a binding site for the nuclear transcriptional repressor CTCF (CCCTC-binding factor) (
34), which are indicative of the ability to enhance the expression of
NET. Interestingly, rs2242446 is also in high linkage disequilibrium (linkage disequilibrium r
2=0.97) with the variant rs28386840 (T-3081A) polymorphism, which is downstream of
NET (2.7 kb). Given their high linkage disequilibrium, this suggests that rs2242446 and rs28386840 may serve as proxy indicators of the functional effect either variant would have on
NET expression. In vitro functional evidence has shown that the wild-type rs28386840 T allele significantly reduces
NET promoter function, resulting in decreased expression (
36). Individuals with the rs28386840 T/T show significantly lower
NET expression in the hippocampus (p=0.013), which is similar to the lower expression seen in those with the rs2242446 C/C genotypes (see Figure S3B and S3A in the
online data supplement) (
35). Given the lower neural expression of
NET and observed better response with C/C genotypes, further investigations are warranted to explore the functional consequences of rs2242446 in
NET gene expression and venlafaxine action.
Our findings should be placed in the context of previous studies: There have been two other studies, in younger individuals, that examined the
NET rs2242446 polymorphism in relation to antidepressant response (
11,
37). In an Australian sample of European ancestry, Singh et al. (
37) reported that individuals with a history of childhood abuse carrying the C allele of the rs2242446 marker were more likely to respond to venlafaxine or escitalopram treatment (N=38). Although these findings are consistent with our results, the sample used by Singh et al. was small and the authors did not conduct genotypic analysis—rather, allelic analysis was conducted comparing C allele carriers (N=29) and T/T genotype carriers (N=9). In a study of Japanese patients with major depression treated with the SNRI milnacipran (N=96), analyses indicated that the rs2242446 T allele was associated with better response, but no genotypic association was detected (
11). This negative finding with respect to genotype is likely due to the smaller sample size, differences in age and in population investigated (i.e., European versus Asian ancestries), and differential affinities for the norepinephrine transporter between venlafaxine and milnacipran. In sum, findings from three studies support the association of rs2242446 with SNRI response. However, the effects may be modulated by additional factors, such as antidepressant type, patient ethnicity, early childhood adverse events, and age, which should be further dissected.
We also observed a nominal association between
HTR2A variant rs6311 (A-1438G) and percentage change in MADRS score across in the total (mixed ancestry) sample and the ethnic subsamples. However, the association was seen only when we adjusted for covariates and not independently with genotype. Variant rs6311 is an upstream (308 base pairs) promoter variant of
HTR2A that may initiate gene transcription. Although functional evidence warrants further investigation of rs6311, a meta-analysis showed no significant association with rs6311 and remission with various antidepressants across 10 studies (
38). Our results add to previous findings (
7,
8,
10,
11) suggesting a minor role for
HTR2A rs6311 in contributing to remission during treatment with venlafaxine.
Overall, our multivariate logistic regression models were able to account for about 25% of the initial variance, suggesting that treatment response may be a complex and polygenic phenotype involving other genes that may also affect venlafaxine metabolism (e.g., CYP2D6) or other mechanisms, such as epigenetic regulation. Nonetheless, our post hoc power analyses for genetic studies revealed that we had a 74% power to detect a multivariate odds ratio of 1.67 or a partial eta squared of 0.03 for rs2242446 with a major allele frequency of 0.74.