Alcohol abuse and dependence (alcoholism) are multifactorial, polygenic disorders involving complex gene-gene and gene-environment interactions. To date, the genes with the strongest associations with alcoholism are those that encode the major enzymes involved in alcohol metabolism, alcohol dehydrogenase and aldehyde dehydrogenase. Two alcohol dehydrogenase genes (
ADH2 and
ADH3 on chromosome 4) and one aldehyde dehydrogenase gene (
ALDH2 on chromosome 12) exhibit functional polymorphisms that influence the rates of conversion of alcohol to acetaldehyde and acetaldehyde to acetate
(1,
2). In humans, three polymorphisms related to alcohol conversion have been identified for
ADH2 (*1, *2, *3), two for
ADH3 (*1, *2), and two for
ALDH2 (*1, *2).
The
ALDH2*2 polymorphism, which is prevalent in Asian populations but extremely rare in non-Asians, has the strongest protective association with alcohol dependence. Asians who are homozygous for
ALDH2*2 have almost zero risk, whereas heterozygotes are about one-third as likely to be alcoholic, compared to those without this allele
(3–
13). According to the main hypothesis about the mechanism underlying this association, the isoenzyme encoded by the
ALDH2*2 allele leads to impaired conversion of acetaldehyde to acetate, causing elevated levels of acetaldehyde
(14–
19), greater sensitivity to alcohol
(16,
20,
21), and lower levels of alcohol consumption
(10,
22–25).
The
ADH2*2 polymorphism has also has been associated with lower rates of alcoholism. This relationship has been found in Asians, after the analyses controlled for the presence of
ALDH2*2
(4–
13,
26,
27), as well as in Caucasians
(28,
29).
ADH2*2 is highly prevalent among Asians and infrequent in most Caucasians except for individuals of Jewish descent
(30,
31). A meta-analysis indicated that individuals with
ADH2*2 are about one-third as likely to be alcoholic, compared to those without this allele
(32). A less common polymorphism of
ADH2, the
ADH2*3 allele, is prevalent in African Americans
(33,
34) and has also been identified, although in low prevalence, among South Africans of mixed ancestry
(35) and Native American Mission Indians of mixed but no known African ancestry
(36).
ADH2*3 has been associated with a negative family history of alcoholism in a pilot study of African Americans, but no associations for alcohol abuse and dependence were found, most likely because of the small number of subjects in the study and the study’s low statistical power
(37).
The
ADH3*1 polymorphism is prevalent in Asians, Caucasians, and African Americans. A few studies have reported that
ADH3*1 might also be associated with lower risk for alcohol dependence
(6,
7,
11). Recent investigations, however, have found that the observed differences in the allele distribution of
ADH3 between alcoholics and comparison subjects can be accounted for by a linkage disequilibrium between
ADH3*1 and
ADH2*2
(4,
28,
38). The
ADH2 and
ADH3 genes are located in tandem on chromosome 4
(39,
40), and polymorphisms at these two loci do not occur independently. Thus, the relationships of
ADH2*2 and
ADH3*1 with lower risk for alcohol dependence appear to be associated.
Method
A total of 340 (213 female participants) Mission Indian adults between the ages of 18 and 73 years who were of mixed heritage but were at least one-sixteenth Native American were recruited from reservations in southern California. An accurate assessment of each participant’s ancestry was not available; however, it was possible to categorize a subgroup (N=246, 72%) according to whether their heritage was <50% (N=123) or ≥50% (N=123) Native American.
Participants were recruited by means of flyers and by word-of-mouth within the Mission Indian community, which consists of approximately 3,000 individuals living on six geographically contiguous reservations. After complete description of the study to subjects, written informed consent was obtained. Each participant completed an interview with the Semi-Structured Assessment for the Genetics of Alcoholism
(45–
47), which was used to make a lifetime diagnosis of alcohol dependence according to DSM-III-R criteria and to assess the maximum number of drinks ever consumed in a 24-hour period. A drink was defined as approximately 9 g of absolute alcohol (e.g., 12 ounces of beer, 4 ounces of wine, or a single shot of hard alcohol), and a list of alcohol equivalents was provided to each participant.
In addition, a blood sample was collected from each participant. Samples were sent to Indiana University for genotyping at the
ALDH2,
ADH2, and
ADH3 loci by using polymerase chain reaction of DNA and allele-specific oligonucleotide probes
(48,
49). Logistic regression analysis and case-control odds ratio calculations were used to examine associations between
ADH2 and
ADH3 allele distributions and alcohol dependence. Multiple regression analysis was used to examine associations between
ADH2 and
ADH3 alleles and the continuous variable, maximum number of drinks. For the logistic regression and multiple regression analyses, gender was first entered as a covariate. Three alleles (
ADH2*2,
ADH2*3, and
ADH3*1) were coded 0, 1, or 2, respectively, and next entered simultaneously into each analysis. The reference group, therefore, consisted of subjects with the
ADH2*1/*1 and
ADH3*2/*2 genotypes. These codings assume linear relationships between each allele and alcohol-related behavior, which is supported by previous association studies of these genes and their hypothesized mechanisms of action, although other models are also possible
(4,
50).
Results
A total of 203 participants (60%) met the criteria for a lifetime DSM-III-R diagnosis of alcohol dependence. Men were significantly more likely to be alcohol dependent (N=91, 72%) than women (N=112, 53%) (χ2=12.03, df=1, p≤0.001).
ALDH2 genotyping indicated that all participants had the ALDH2*1/*1 genotype, thus the ALDH2*1 allele frequency was 1.0. ADH2 genotyping indicated that 299 subjects had the ADH2*1/*1 genotype, 20 had ADH2*1/*2, two had ADH2*2/*2, 17 had ADH2*1/*3, and two had ADH2*2/*3. Thus, the ADH2*1 allele frequency was 0.93, the ADH2*2 allele frequency was 0.04, and the ADH2*3 allele frequency was 0.03. ADH3 genotyping indicated that 157 subjects had the ADH3*1/*1 genotype, 131 had ADH3*1/*2, and 52 had ADH3*2/*2. Thus, the ADH3*1 allele frequency was 0.65 and the ADH3*2 allele frequency was 0.35. Men and women did not differ significantly in their allele distributions of ADH2 (χ2=0.09, df=2, p=0.96) or ADH3 (χ2=0.93, df=1, p=0.34).
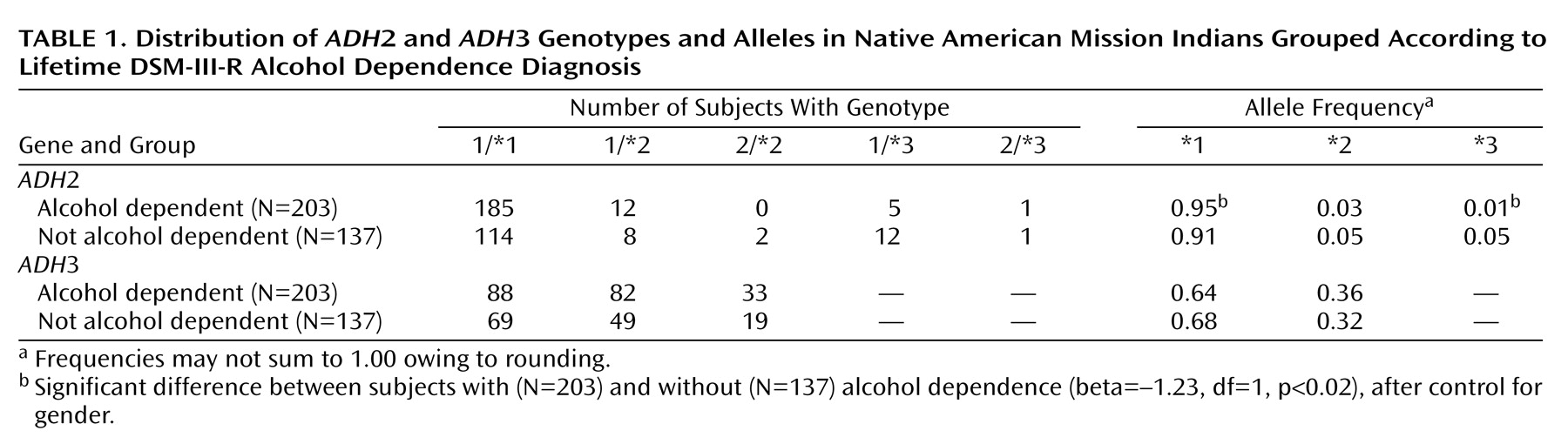
After covarying for gender (beta=–0.85, df=1, p≤0.0001), logistic regression analysis revealed a significant association between
ADH2*3 and alcohol dependence (beta=–1.23, df=1, p<0.02), but
ADH2*2 (beta=–0.28, df=1, p=0.48) and
ADH3*1 (beta=–0.17, df=1, p=0.30) were not significantly associated with alcohol dependence. Case-control odds ratio calculations, which controlled for gender, indicated that participants with alcohol dependence were significantly less likely to have the
ADH2*3 allele (odds ratio=0.28, 95% confidence interval [CI]=0.10–0.77, p<0.02) and significantly more likely to have the
ADH2*1 allele (odds ratio=2.00, 95% CI=1.10–3.65, p<0.03), compared to those who were not alcohol dependent. No significant differences were found between alcohol-dependent and non-alcohol-dependent participants in the allele distributions of
ADH2*2 (odds ratio=0.70, 95% CI=0.33–1.49, p=0.35),
ADH3*1 (odds ratio=0.81, 95% CI=0.60–1.10, p=0.17), or
ADH3*2 (odds ratio=1.24, 95% CI=0.91–1.70, p=0.17), after control for gender.
Table 1 shows the
ADH2 and
ADH3 genotype and allele distributions grouped according to lifetime diagnosis of alcohol dependence.
For the variable maximum number of drinks ever consumed in a 24-hour period, data were missing for two non-alcohol-dependent male subjects, one with the
ADH2*1/*1 and
ADH3*1/*2 genotypes and one with the
ADH2*2/*2 and
ADH3*1/*1 genotypes, and these subjects were excluded from the analyses. The remaining 338 participants reported consuming an average maximum of 26.9 drinks (SD=23.2, range=1 to 192). Analyses revealed that this variable was positively skewed. A natural logarithm (log) transformation was used to normalize the distribution for all analyses, but values from untransformed data are presented. Consistent with previous research
(44), men reported a significantly higher maximum number of drinks (mean=34.1, SD=26.9, N=125) than women (mean=22.7, SD=19.6, N=213) (F=20.03, df=1, 336, p≤0.0001), and participants with a diagnosis of alcohol dependence reported a significantly higher maximum number of drinks (mean=34.0, SD=24.6, N=203) than those without alcohol dependence (mean=16.2, SD=15.8, N=135) (F=125.86, df=1, 336, p≤0.0001).
After covarying for gender (F=19.26, df=1, 333, p≤0.0001), multiple regression analysis revealed a significant association between ADH2*3 and the log of maximum number of drinks (F=5.48, df=1, 331, p=0.003), but
ADH2*2 (F=0.18, df=1, 333, p=0.59) and
ADH3*1 (F=0.17, df=1, 333, p=0.60) were not significantly associated with the log of maximum number of drinks.
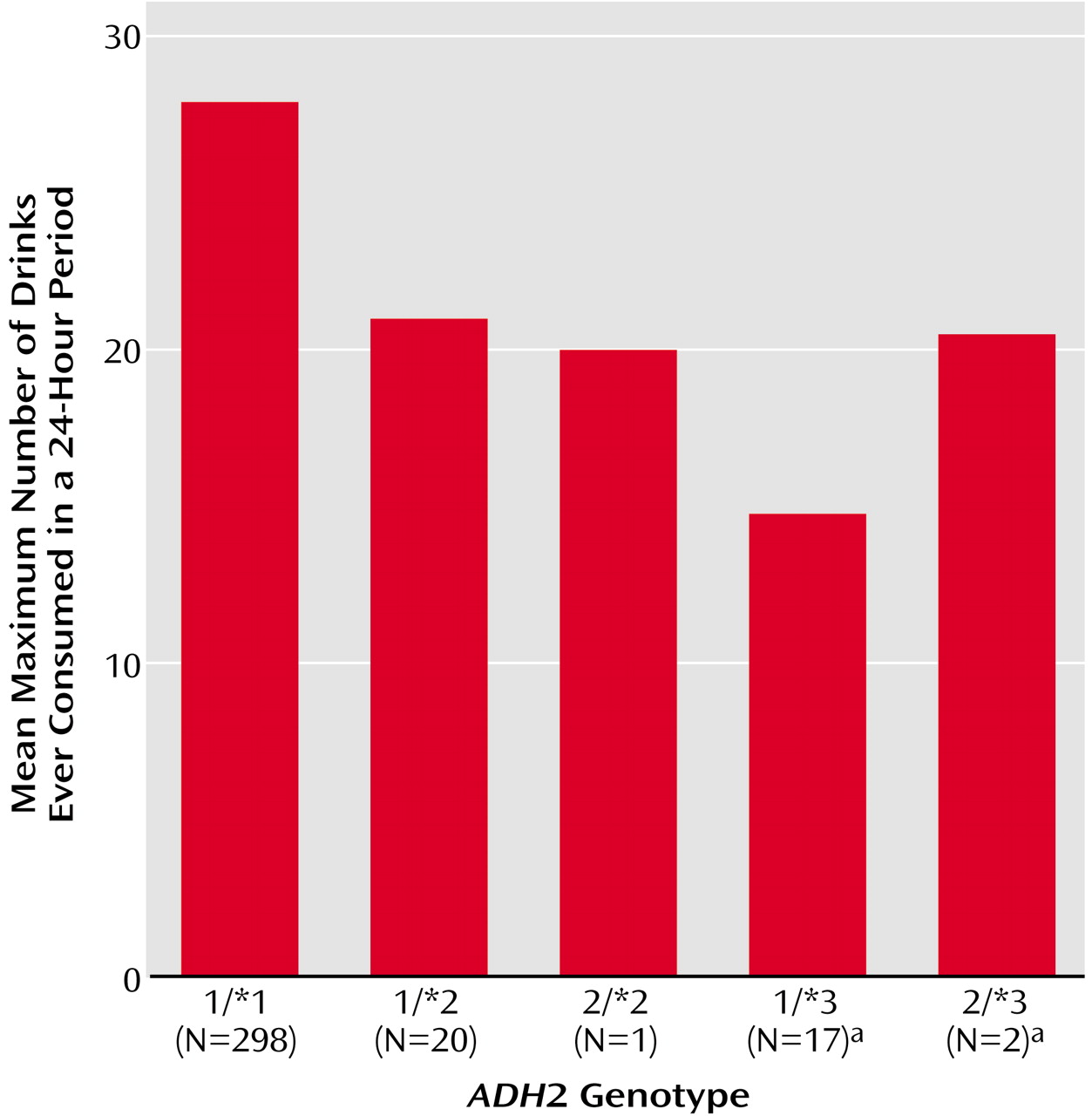
Figure 1 displays the maximum number of drinks ever consumed in a 24-hour period by subjects grouped according to ADH2 genotype. Participants with an
ADH2*3 allele reported a lower maximum number of drinks (mean=15.4, SD=10.2, N=19), compared to those without an
ADH2*3 allele (mean=27.6, SD=23.6, N=319).
Because of the potential for population stratification, additional analyses were conducted on data for the subgroup of participants who were categorized as having either <50% (N=123) or ≥50% (N=123) Native American heritage. No significant associations were found between Native American heritage and the allele distributions of ADH2 (χ2=3.27, df=2, p=0.20) or ADH3 (χ2=0.01, df=1, p=0.92). Mission Indians with ≥50% Native American heritage were significantly more likely to be alcohol dependent than those with <50% Native American heritage (70% versus 54%; χ2=6.89, df=1, p=0.009). No significant association was found between Native American heritage and the log of the maximum number of drinks ever consumed (F=0.29, df=1, 242, p=0.59); the mean maximum number of drinks was 29.9 (SD=25.2) for those with ≥50% Native American heritage and 28.2 (SD=25.2) for those with <50% Native American heritage.
Discussion
The Mission Indians evaluated in this study had a high rate of alcohol dependence in both men (72%) and women (53%), which is consistent with investigations of other Native American tribes
(51–
53). This study also replicated previous work from our laboratory that found a low prevalence of
ADH2*2 and
ADH2*3, two alleles that have been associated with alcohol-related behavior in other populations. In a prior report on 95 Mission Indian men with at least one-eighth Native American ancestry, the prevalence of
ADH2*2 was 0.01 and the prevalence of
ADH2*3 was 0.06
(36), compared with prevalences of 0.04 and 0.03, respectively, for these alleles in the current study’s larger group of participants with a greater degree of admixture.
A protective association of
ADH2*3 with alcohol dependence was also found in this study. Mission Indians with an
ADH2*3 allele were about one-third less likely than those without this allele to have a lifetime diagnosis of alcohol dependence (odds ratio=0.28). This reduced risk for alcohol dependence is approximately equal to that associated with having either one
ALDH2*2 or one
ADH2*2 allele in previous studies of other ethnic groups
(3–
13,
26–
29,
32). In addition, individuals with
ADH2*3 reported a maximum number of drinks ever consumed in a 24-hour period that was almost half that reported by those without this allele. As well as being related to a diagnosis of alcohol dependence, this endophenotype has been found to have a heritability of approximately 50% in a study of adult Australian twins (A.C. Heath, personal communication, 2002) and was related to the
ALDH2*2 allele in a study of Asian Americans
(54). The present findings suggest that the investigation of genetic associations with the presence or absence of disorders can be enhanced by also evaluating related behaviors measured as continuous variables, where relationships may be more easily detected.
Moreover, these results are consistent with genetic linkage studies showing protective associations for alcohol-related behavior on chromosome 4. Genome scans of family members from the Collaborative Study on the Genetics of Alcoholism found evidence suggestive of protection against alcohol dependence
(55), as well as a lower number of maximum drinks ever consumed in a 24-hour period
(44), on the area of chromosome 4 that includes the
ADH2 and
ADH3 genes.
ADH3 genotyping indicated that the maximum number of drinks was not associated with variation at this locus
(44), but
ADH2 polymorphisms were not evaluated. A similar association with alcohol dependence on chromosome 4 was reported from a genome screen of Native Americans
(56). Results from the present study suggest
ADH2 polymorphisms may account for these findings.
ADH2*2,
ADH3*1, and
ADH3*2 were not associated with alcohol-related behavior in this study. Although Mission Indians with the
ADH2*2 allele were less likely to be alcohol dependent and reported a lower maximum number of drinks in a 24-hour period (
Figure 1) than individuals with the
ADH2*1/*1 genotype, the differences were not statistically significant. This null finding may be related to the low prevalence of the
ADH2*2 allele in this population, although a similar
ADH2*2 prevalence is found in non-Jewish Caucasians, for whom a protective association with alcohol dependence has been observed
(28,
29).
Studies of children with fetal alcohol syndrome have found protective associations of
ADH2*2 in South Africans of mixed ancestry
(35) and of
ADH2*3 in African Americans
(34,
43,
57). It has been hypothesized that faster alcohol metabolism leading to a more rapid production of acetaldehyde, increased sensitivity to alcohol, and lower levels of alcohol consumption is the mechanism by which these alleles protect against both fetal alcohol syndrome and alcohol dependence.
ADH2*3 has been associated with faster alcohol metabolism in African Americans
(33) and a nonsignificant trend was observed in a small group of Mission Indian men
(42), but other studies have not found significant associations between
ADH2*2 and alcohol metabolism
(19,
58) or alcohol sensitivity
(59). In addition, there is no direct evidence that faster alcohol metabolism leads to greater production of acetaldehyde. Results from this study support the hypothesis that the protective association between
ADH2*3 and alcohol dependence is mediated, in part, by lower levels of alcohol consumption. Additional research is needed to determine if
ADH2*3 is also associated with elevated levels of acetaldehyde or greater sensitivity to alcohol.
It is also possible that the association of ADH2*3 with alcohol-related behavior in this group of subjects may not be a genuine causal effect, but may be related to either linkage disequilibrium with a nearby locus or population stratification because of racial admixture. Although an attempt was made to address concerns about heterogeneity, complete data on all participants’ Native American heritage were not available. Therefore, it will be important to determine the generalizability of the present findings for other Native American and non–Native American populations, particularly for African Americans, among whom ADH2*3 is prevalent.
In conclusion, this study provides evidence that the ADH2*3 allele is associated with lower rates of alcohol dependence and lower rates of heavy drinking. These findings also highlight the utility of evaluating protective factors in populations with high rates of alcohol dependence. Although the prevalence of ADH2*3 was low in this group of Mission Indians, the high prevalence of alcohol dependence and large variability in drinking behavior in this Native American tribe made it possible to detect significant associations between this genetic polymorphism and alcohol-related behavior.