Pharmacogenetics research focuses on the identification of genetic variants that predict which individuals may optimally benefit from antipsychotic treatment (
5). Variants in genes that code for neurotransmitter receptors have been the primary targets, including multiple loci in the dopamine and serotonin receptor systems. However, there remain surprisingly few studies on the relationship between the most obvious candidate gene,
DRD2, and antipsychotic drug response. Several lines of evidence suggest that the dopamine type 2 (D
2) receptor plays a critical role in antipsychotic drug action. Earlier studies showed that antipsychotic clinical potency is highly correlated with the binding affinity to a particular type of dopamine receptor (
6), which was later found to be the D
2 receptor (
7,
8). Recent functional imaging studies suggest that binding to the D
2 receptor by antipsychotic agents may be "necessary and sufficient" for antipsychotic efficacy (
9). Finally, all known antipsychotic drugs bind to the D
2 receptor, and drugs that have targeted non-D
2 receptors without at least some element of D
2 blockade have failed to treat schizophrenia effectively (
8,
10).
As we describe in detail, relevant studies in the literature have utilized a variety of designs, trial durations, symptom measures, and response criteria. Consequently, we developed a systematic and consistent methodology to harmonize the phenotypes reported, contacting the original investigators when re-evaluating raw data was necessary. Additionally, while multiple
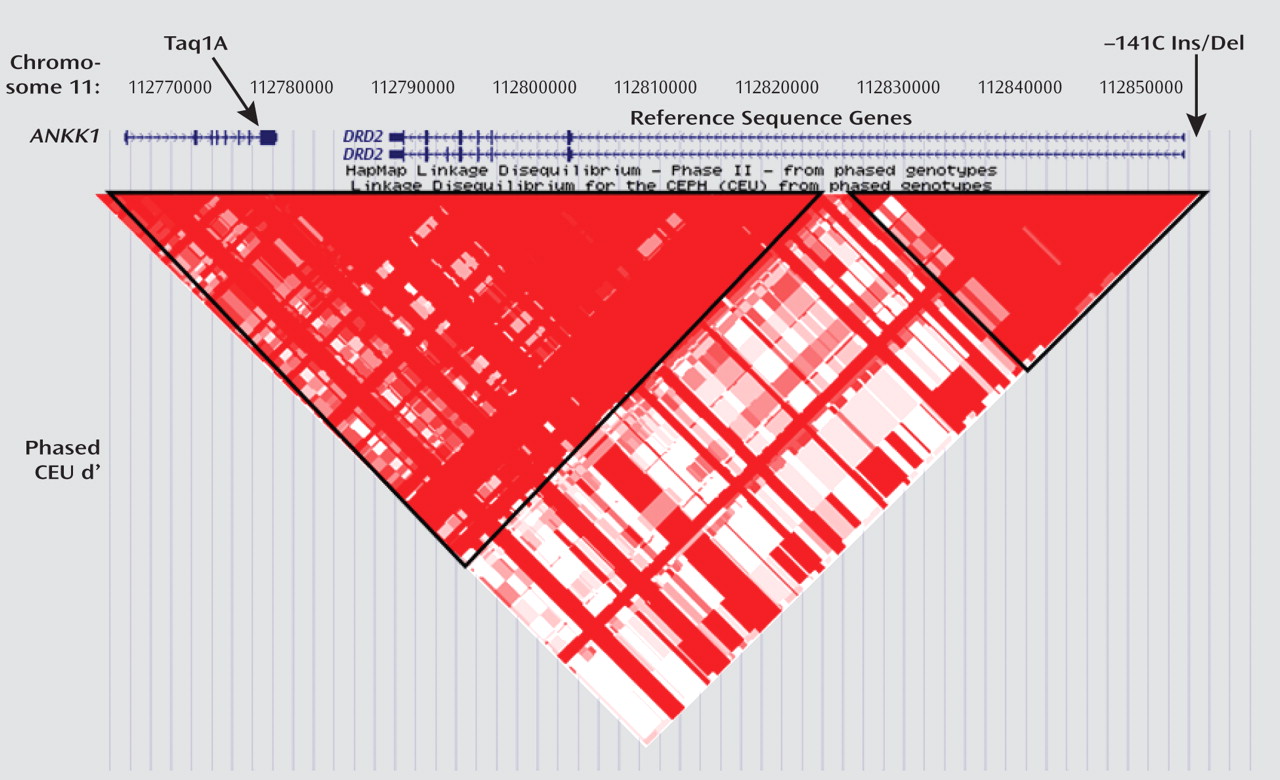
DRD2 SNPs have been studied, including Taq1B (
14,
15), Taq1D (
15,
16), T939C (
14), S311C (
17), and C957T (
16,
18), most of these SNPs were reported in only one or two studies, with the exception of the Taq1A and –141C Ins/Del variants. Finally, studies to date have included patients across all phases of the illness, ranging from first-episode schizophrenia patients, with no or limited prior exposure to antipsychotic drugs, to clozapine-treated patients, with poor prior antipsychotic drug responses and lengthy medication histories. Since antipsychotic drug exposure has been demonstrated to alter the expression of multiple CNS receptors (
19), including the D
2 receptor, this factor may introduce additional variance into studies of genetic sources of variability. Therefore, we conducted exploratory analyses to investigate whether studies examining first-episode cohorts yielded stronger results than those consisting primarily of chronically ill subjects.
Results
To conduct our meta-analysis on the relationship between
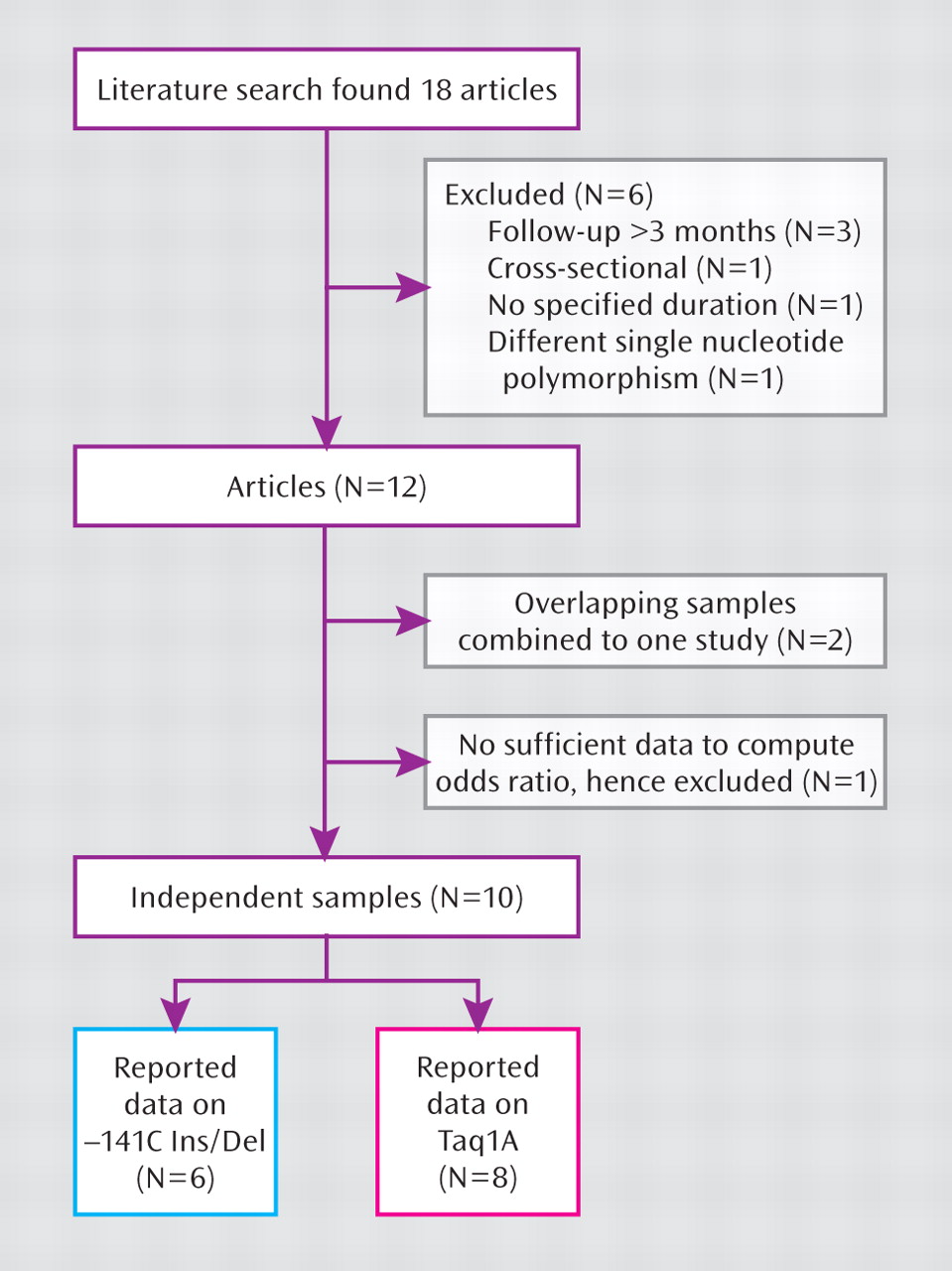
DRD2 gene variation and antipsychotic drug response, we searched for literature available up to December 31, 2008, which yielded 18 published articles. Of these, six articles were not included because three (
15,
16,
34) only reported long-term outcomes >3 months from baseline to follow-up evaluation, one (
35) was cross-sectional, one (
36) did not state the duration of follow-up evaluation or include a standardized rating scale such as the BPRS, PANSS, or CGI, and one (
17) contained no data on Taq1A or –141C Ins/Del polymorphisms. Two articles published by the same research group (
37,
38) contained overlapping data and therefore were assessed as one study, subsequent to the authors providing the additional data needed to compute the categorical outcome of clinical response. Finally, in one other study (
39), we were unable to obtain sufficient information to calculate response rates, and therefore we did not include data from that study.
A total of 10 independent studies (total sample size: N=889) met criteria for inclusion in the present meta-analysis.
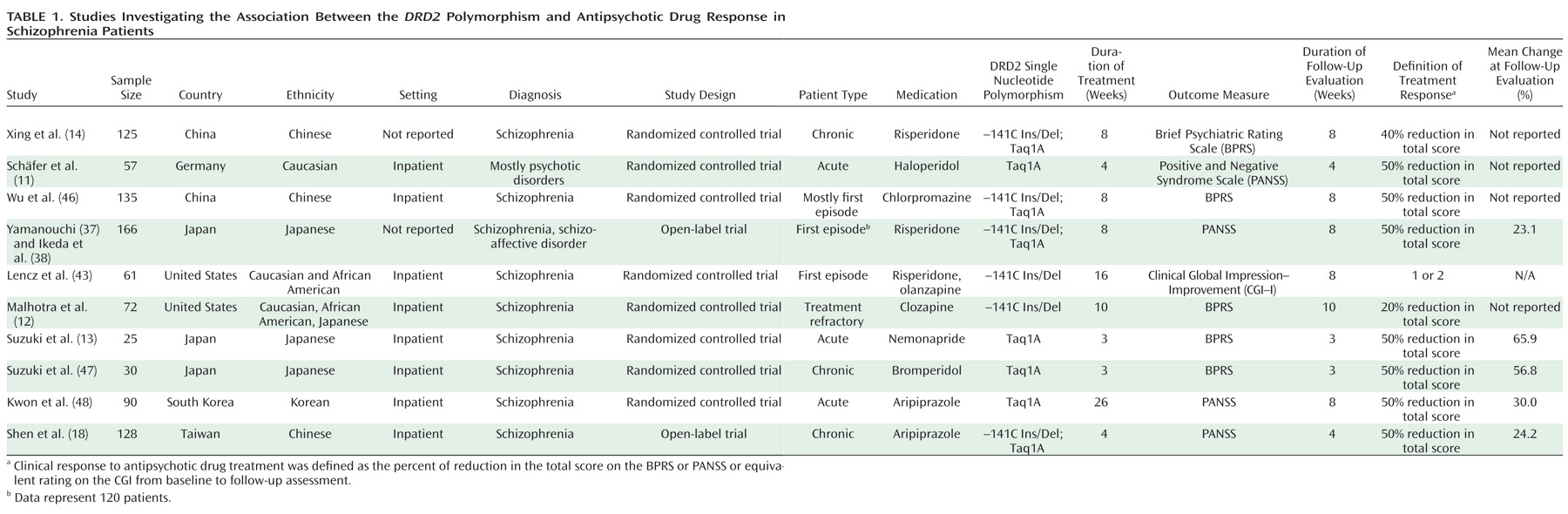
Figure 2 illustrates the literature search process. The clinical characteristics of each study are summarized in
Table 1. Six studies reported outcomes conditioned on the –141C Ins/Del SNP (total sample size: N=687), and eight studies reported outcomes conditioned on the Taq1A SNP (total sample size: N=748). In addition, six studies reported continuous outcomes, but the authors generously provided the additional data needed to compute odds ratios.
–141C Ins/Del Polymorphism and Antipsychotic Drug Response
As mentioned earlier, six studies that met inclusion criteria reported results on the –141C Ins/Del polymorphism, with a total sample size of 687 patients.
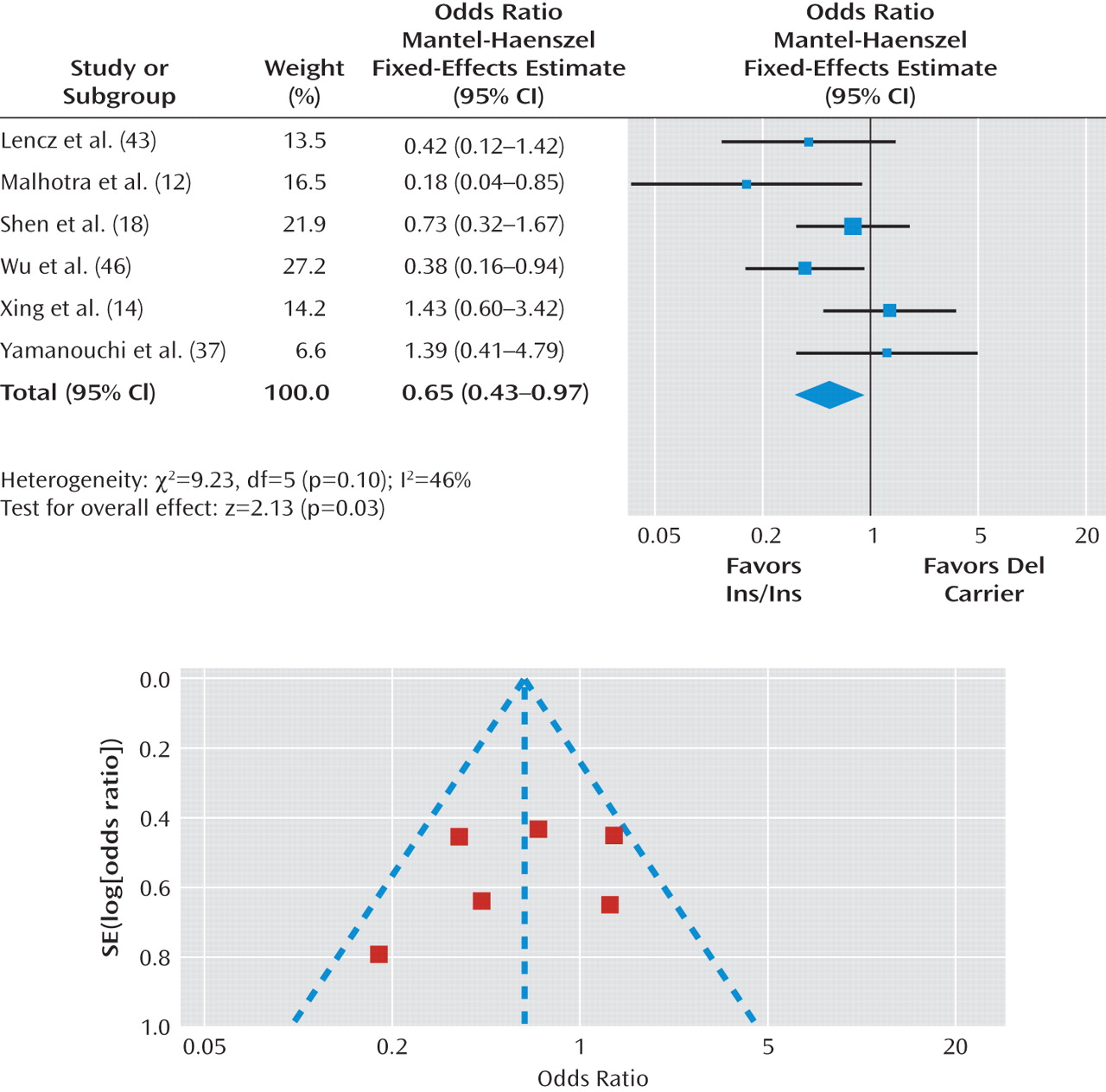
Figure 3 presents odds ratios for the individual studies and the pooled analyses in different genotype groups. There was a significant difference in the response rate between the Del carrier and Ins/Ins genotype groups (pooled odds ratio=0.65, 95% CI=0.43 to 0.97, p=0.03), indicating that Del carriers tend to have less favorable antipsychotic drug responses than individuals with the Ins/Ins genotype. The chi-square test assessing heterogeneity did not reveal significance (χ
2=9.23, df=5, p=0.10; I
2=46%). To deal with potentially undetected heterogeneity across samples, we conducted a sensitivity analysis that excluded the study with the largest effect size (
12) and the study with the smallest effect size (
14). Another reason to exclude these two studies was that they may be different from other studies because the 50% reduction of the BPRS or PANSS total score was not used to define clinical response. For the sensitivity analysis, I2 was changed from 46% to 10% and the chi-square test for heterogeneity revealed a decrease from 9.23 to 3.33, which was nonsignificant, with a change in the p value from 0.10 to 0.34. The pooled odds ratio became 0.60, with a 95% CI range of 0.38–0.97 and p value of 0.04.
In a post hoc analysis, we restricted our investigation to studies consisting of patients with first-episode schizophrenia (total sample size: N=316). The pooled odds ratio for Del carrier patients relative to patients with the Ins/Ins genotype was 0.53 (95% CI=0.28 to 0.99, p=0.05), demonstrating poorer clinical response in Del carriers. In contrast, for studies that did not include first-episode schizophrenia patients (total sample size: N=371), we obtained a pooled odds ratio of 0.75 (95% CI=0.44 to 1.27, p=0.23) (see Figure 1 in the data supplement accompanying the online version of this article).
As seen in Figure 3, funnel plot analysis did not demonstrate evidence of publication bias. The Duval and Tweedie (
32) trim and fill method indicated that it was not necessary to trim any existing study and fill any additional unpublished study. In addition, Egger's test (
33) also revealed no evidence of publication bias (beta=0.79, 95% CI=–3.05 to 4.63, p=0.60).
Taq1A Polymorphism and Antipsychotic Drug Response
As previously discussed, eight studies assessed the Taq1A polymorphism and antipsychotic response, with a total sample size of 748 patients. Odds ratios for the individual studies and the pooled analyses in different genotype groups are shown in
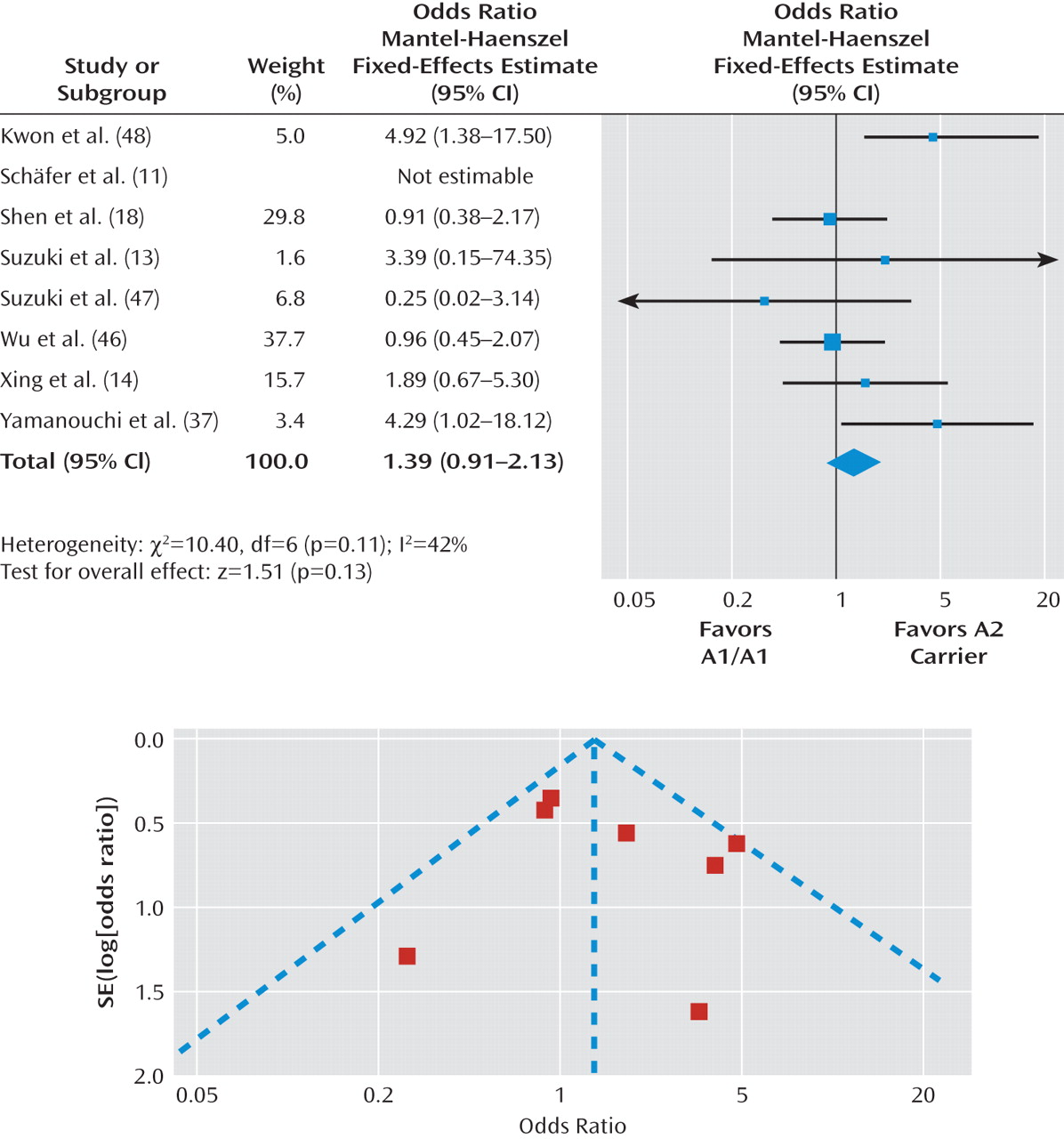
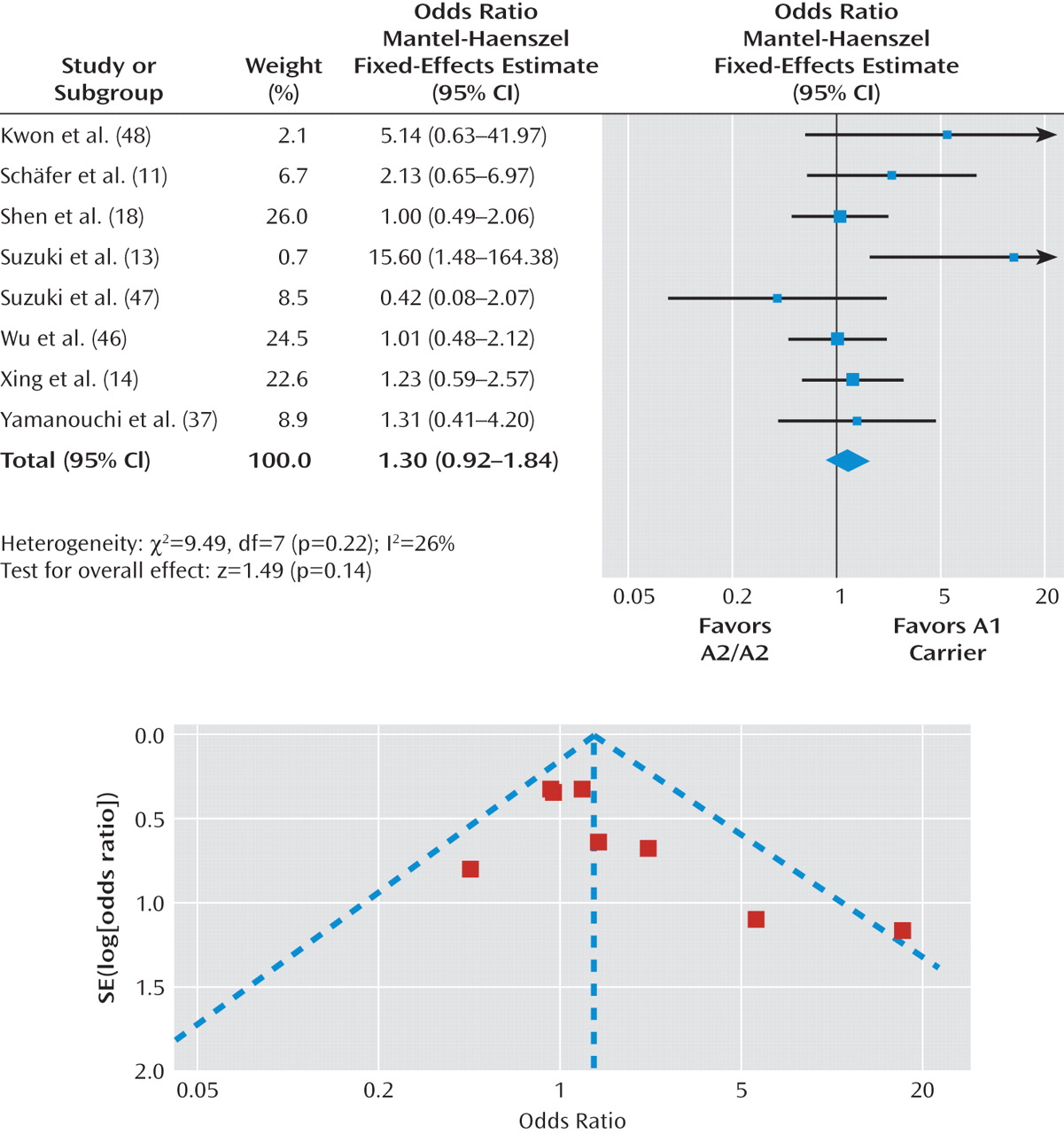
Figure 4 and
Figure 5. There was no significant difference in the treatment response rate among individuals with the A1/A1 genotype relative to A2 allele carriers (pooled odds ratio=1.39, p=0.13 [Figure 4]) or A1 allele carriers relative to individuals with the A2/A2 genotype (pooled odds ratio=1.30, p=0.14 [Figure 5]). Further, there was no significant heterogeneity across studies in these two comparisons (A1/A1 genotype relative to A2 allele carriers: χ
2=10.40, p=0.11; I
2=42%; A1 allele carriers relative to individuals with the A2/A2 genotype: χ
2=9.49, p=0.22; I
2=26%).
The Duval and Tweedie trim and fill analysis showed that it was necessary to fill an additional unpublished study for both the A1/A1 genotype versus A2 allele carrier comparison (pooled odds ratio=1.24, p=0.30) and A1 allele carrier versus A2/A2 genotype comparison (pooled odds ratio=1.17, p=0.39). In contrast, Egger's test revealed no evidence of publication bias for either comparison (A1/A1 genotype versus A2 allele carrier: beta=-0.07, p=0.93; A1 allele carrier versus A2/A2 genotype: beta=-0.60, p=0.26). In summary, the evidence regarding publication bias for the Taq1A polymorphism was inconsistent. Even if we were able to eliminate publication bias, it appears that the association between the Taq1A polymorphism and antipsychotic drug response would still not be significant.
Discussion
In order to assess the relationship between DRD2 genetic variation and antipsychotic drug response, we conducted the first meta-analysis of the –141C Ins/Del and Taq1A polymorphisms, two commonly studied DRD2 SNPs, and clinical response to antipsychotic drug treatment. The primary result was that the –141C Ins/Del polymorphism significantly influenced antipsychotic drug response (total sample size: N=687), whereas we were not able to detect a relationship between clinical response and the Taq1A variant.
These data are consistent with prior research indicating an important role for the D
2 receptor in antipsychotic drug response. Antipsychotic clinical potency is highly correlated with the binding affinity to the D
2 receptor (
6–
8); D
2 receptor occupancy by antipsychotic agents has been demonstrated to occur with all antipsychotic agents (
9); and drugs targeting other receptor sites without D
2 blockade have not yet been successfully developed as antipsychotics (
8). To our knowledge, this is the first meta-analysis in pharmacogenetics to demonstrate the importance of
DRD2 genetic variation in antipsychotic drug response.
Of note, we observed a significant genotype-phenotype relationship in patients with first-episode schizophrenia. This may be the result of limited or lack of prior exposure to antipsychotic drug treatment in these patients. Differential amounts of prior drug exposure, as commonly observed in chronically ill samples, could result in considerable variation in levels of dopamine receptor up-regulation (
40,
41) and potentially mask subtle genetic effects on dopamine receptor availability (
42) that could mediate antipsychotic response. However, other factors, including greater drug response rates in first-episode patients, should be considered as well as the limitation that studies on first-episode patients are less common than studies on chronically ill patients.
There was no significant association between the Taq1A polymorphism and antipsychotic drug response in the eight studies reporting outcomes conditioned on the Taq1A SNP, with a total sample size of 748 patients. Although the Taq1A polymorphism has been found to be associated with drug response in several studies (
11,
18,
38), it is not clear how it is related to the
DRD2 gene, and it is actually located in a noncoding region of the
DRD2 locus. In contrast, we did find a significant association between the –141C Ins/Del polymorphism and antipsychotic drug response. This may be because this SNP is located in the 5′ promoter region of
DRD2, where it may influence modulation of transcriptional activities (
23) and D
2 receptor density (
42). Interestingly, another
DRD2 SNP, A-241G, which is also located in the promoter region, has been associated with antipsychotic drug response (
43).
Although sample size limitations do not provide us with an opportunity to conduct drug-specific analysis, it is not unexpected that DRD2 variation might influence clinical response to all antipsychotics. First, all antipsychotic drugs bind potently to the D2 receptors. Second, there are few data to suggest that any one antipsychotic has markedly improved efficacy over another, and similar response rates suggest phenotypic overlap and provide the rationale for the grouping of individual drug responses for analysis. Third, and perhaps most importantly, each of the antipsychotic drugs was specifically developed because of the common mechanism of action of antagonism of D2 receptors, and therefore a common effect of DRD2 variation across these drugs seems highly plausible. Nevertheless, the development of drugs with antipsychotic efficacy that does not act at the D2 receptor will be needed to empirically assess this issue.
There are several limitations of this study. First, odds ratio was used as the effect-size measure. Because this requires dichotomizing a continuous measure of either BPRS or PANSS scores, statistical power may have been diminished and it may explain why some studies reported significant findings of an association between DRD2 and antipsychotic drug response while the odds ratios were not individually significant. Therefore, meta-analysis of odds ratios may lack some sensitivity to detect small effect sizes. This is consistent with an exploratory sensitivity analysis using a random-effect model (see Figure 2 in the online data supplement), which produced a less robust p value than the fixed-effect model. Nevertheless, categorical response, instead of incremental differences in scores on the BPRS or PANSS, may be more meaningful from a clinical perspective. To clarify the clinical relevance of DRD2 genetic variations, it may be necessary to use an even more clinically meaningful outcome measure, such as the number of days to hospital discharge following acute treatment or assessments of functional disability.
Second, variation in the antipsychotic drugs administered in the studies we analyzed limited the possibility of examining the association of
DRD2 with any specific drug. In these studies, multiple antipsychotic drugs were utilized, including typical agents such as chlorpromazine and haloperidol and atypical drugs such as clozapine, risperidone, olanzapine, and aripiprazole. Although all of these drugs act on the D
2 receptor, they exhibit different affinity profiles for many of the candidate receptors (
44), making direct comparisons more complex. For example, non-D
2 receptors, such as D
3, D
4, and serotonin 5-HT
2A, may also be important in antipsychotic drug action (
44) as well as new mechanisms of action, such as metabotropic glutamate receptor 2 and receptor 3 stimulation (
45). Additionally, it should be noted that the studies included patients from several different ethnic groups, with an overrepresentation of Asian patients (e.g., Chinese, Korean, and Japanese populations) and an underrepresentation of individuals of African descent. Since allele frequencies may vary considerably between ethnic groups, careful consideration of the potential effect of population genetics on genotypic and phenotypic distribution is warranted, but the limited samples currently available have hampered this effort. Finally, the relatively small number of studies included in this meta-analysis makes it difficult to conduct any meaningful moderator analyses.
As a result of the heterogeneity of medication used, the duration of illness in different samples, and the different racial groups, it is possible that we have underestimated the effect size of the gene-drug response association. Furthermore, none of the studies formally accounted for medication noncompliance, which is prevalent in patients with schizophrenia. Put simply, when a patient does not take the prescribed antipsychotic drug, the measured effect size of gene-drug response association is assessed as zero, whereas the true effect of genotype on the phenotype is perhaps larger. Nevertheless, despite the potential underestimation of effect size produced by these uncontrolled factors, we were still able to detect a significant association between the –141C Ins/Del polymorphism and antipsychotic drug response. Data on the –141C Ins/Del polymorphism from larger studies, such as the Clinical Antipsychotic Trials of Intervention Effectiveness and industry efforts, will be informative and important in further establishing the role of this SNP in antipsychotic drug response.
In summary, our meta-analysis indicates that DRD2 genetic variation is significantly associated with antipsychotic drug response. SNPs in the DRD2 promoter region, such as –141C Ins/Del, may be particularly important in predicting clinical response to antipsychotic drug treatment. Studies with larger cohorts examined with prospective designs may be needed to fully understand the nature of this relationship.