The NOTCH4 protein is a member of the NOTCH family, a set of four type 1 transmembrane proteins that together form a signaling system that plays a key role in cell fate decisions (
13). In addition to being important in regulating neural cell proliferation, neural cell differentiation, and neural cellular growth, the NOTCH4 protein is a crucial contributor in T cell-mediated immune responses (
14,
15). The NOTCH4 protein has also been implicated in endothelial cell dysregulation and vascular inflammation as well as macrophage activation (
16).
NOTCH4 is expressed in endothelial cells and therefore present in various tissues and in the CNS (
14,
17). To further elucidate the potential role of NOTCH4 in schizophrenia and bipolar disorder, we examined
NOTCH4 mRNA expression in the whole blood of patients with schizophrenia, bipolar disorder, and other psychoses, with comparative analyses in healthy comparison subjects. We controlled for several potential confounders and identified expression quantitative trait loci in and around the
NOTCH4 gene that may regulate its expression.
Method
Study Design and Ethics
The Thematically Organized Psychosis Study at the Stavanger University Hospital and Oslo University Hospital, Norway, is a large ongoing study with 1,726 current participants (1,171 patients and 555 healthy comparison subjects). The study sample consisted of patients and comparison subjects included within a particular time period (August 2003–December 2008). The study was approved by the Regional Committee for Medical Research Ethics and the Norwegian Data Inspectorate, and the biobank is approved by the Norwegian Directorate of Health.
Participants
The main inclusion criteria were DSM-IV diagnoses of psychosis disorders, either among the schizophrenia spectrum disorders or the bipolar spectrum disorders, and age between 18 and 65 years. Patients were recruited from in- and outpatient clinics. We did not include patients unless they were clinically stable enough to fully understand the information about the study given to them. All patients gave written informed consent and were made aware that they could withdraw from the study at any time. After completing the protocol, the treating clinician received a full report with somatic and psychiatric assessments of the patient. Exclusion criteria were head injury, neurological disorder, mental retardation, autoimmune and infectious disorders, and malignancies. All patients underwent standardized psychiatric interviews with the Positive and Negative Syndrome Scale (PANSS), the Inventory of Depressive Symptoms, and the Young Mania Rating Scale. All participants were assessed by trained psychiatrists and clinical psychologists using the Structured Clinical Interview for DSM-IV Axis I Disorders (SCID), and they received a diagnosis of either schizophrenia or bipolar spectrum disorder. The type and amount of illicit substances and number of international units of alcohol consumed over the past 2 weeks were recorded. In addition, all participants were screened and diagnosed for illicit substances and alcohol using the E module in the SCID manual.
Patients were divided into groups according to their diagnosis and treated as unitary illnesses. The bipolar disorder group included patients with bipolar I disorder (N=107), bipolar II disorder (N=50), and bipolar disorder not otherwise specified (N=12). The other psychosis group included patients with depressive psychosis (N=22) and psychosis not otherwise specified (N=61). In addition to patients meeting the criteria for schizophrenia (N=175), the schizophrenia group included patients with schizophreniform disorder (N=19) and schizoaffective disorder (N=33). All patients were examined by a physician, and fasting blood samples were drawn in the morning.
Comparison Subjects
A representative age- and sex-matched group of healthy volunteers (N=211) from the same catchment area was randomly selected from the National Population Registry by a computer-based program, and individuals were contacted by a letter of invitation. All comparison subjects were screened for illness using the Primary Care Evaluation of Mental Disorders and interviews about severe psychiatric disorders, drug abuse, and somatic disease.
Exclusion criteria were any history of severe psychiatric disorders (major depression, bipolar disorders, and schizophrenia) in the comparison subjects or in any of their first-degree relatives, substance or alcohol abuse or dependency, medical conditions involving the CNS, and severe head injury. Blood was drawn between 8 a.m. and 7 p.m., but, as opposed to the patient samples, all blood samples from the comparison subjects were not fasting samples. None of the patients or the comparison subjects had any acute infections at the time of blood sampling.
mRNA Analyses
The extraction was done at the same time for patients and comparison subjects and under equal conditions. Details on the RNA isolation, reverse transcription, and real-time polymerase chain reaction are provided in the
data supplement that accompanies the online edition of this article. We investigated potential variations of
NOTCH4 mRNA expression in subpopulations by isolating and analyzing different blood cells in an additional sample of bipolar disorder patients (N=15) and an age- and sex-matched healthy comparison group (N=15).
Genotyping
Only participants with Caucasian ethnicity were genotyped. The number of individuals in each category eligible for the genotype analysis was as follows: bipolar disorder (N=113), schizophrenia (N=144), psychosis not otherwise specified (N=52), and healthy comparison subjects (N=151). Nineteen SNPs in or around
NOTCH4 were genotyped in all patients and comparison subjects with the Affymetrix Genome-Wide Human SNP Array 6.0 (Affymetrix Inc., Santa Clara, Calif.). One additional SNP (rs3131296) was imputed. Ungenotyped markers were imputed by IMPUTE, version 2 (
18) using the Utah sample from the 1000 Genomes Project, with ancestry from northern and western Europe as reference.
Routine Laboratory Analyses
Hemoglobin, C-reactive protein, creatinine, alanine aminotransferase, and serum cortisol were measured using standard laboratory methods.
Statistical Analyses
Statistical analyses testing the association between diagnosis and mRNA level were done using the SPSS software package for Windows, version 18.0 (SPSS, Chicago). Data normality was assessed using the one-sample Kolmogorov-Smirnov test. When using regression models and analysis of variance (ANOVA), all skewed data were log-transformed before the analyses. All analyses were two-tailed with a level of significance of p<0.05. Potential differences between the diagnostic groups and the comparison group were investigated using the chi-square test for categorical variables, the Kruskal-Wallis test for continuous variables, and the Mann-Whitney U test for post hoc analyses. The variation in
NOTCH4 expression across groups was initially investigated with a simple ANOVA model followed by Tukey’s post hoc analysis and adjusted for covariates in an analysis of covariance (ANCOVA) model. We tested for association between 20 SNPs in or around
NOTCH4 and for diagnosis using the allelic test in PLINK (
19). Further details are given in the online
data supplement.
Results
Demographic and clinical variables are summarized in
Table 1. Briefly, patients with bipolar disorder and healthy comparison subjects were slightly older than individuals in the schizophrenia and other psychosis groups. The majority of patients were of European origin (
Table 1). The use of tobacco and cannabis was equally distributed in the three patient groups, but use was significantly higher than it was in the comparison group. Intake of alcohol 2 weeks before blood sample collection varied significantly across all groups. The other psychosis group used the highest amount, followed by the bipolar disorder group, the healthy comparison group, and the schizophrenia group (
Table 1). Body mass index (BMI) was higher in the schizophrenia group relative to the bipolar disorder and healthy comparison groups (
Table 1). The use of antipsychotics and mood stabilizers and the level of PANSS-measured psychotic symptoms varied across the diagnostic groups as expected according to the diagnoses. The schizophrenia group had a higher mean score on mania symptoms as measured by the Young Mania Rating Scale, and the other psychosis group had more depression symptoms than the bipolar disorder group (
Table 1).
NOTCH4 Expression
We found a significant difference in
NOTCH4 mRNA levels across the different diagnostic groups (F=7.1, df=3, 686, p<0.0001). As depicted in
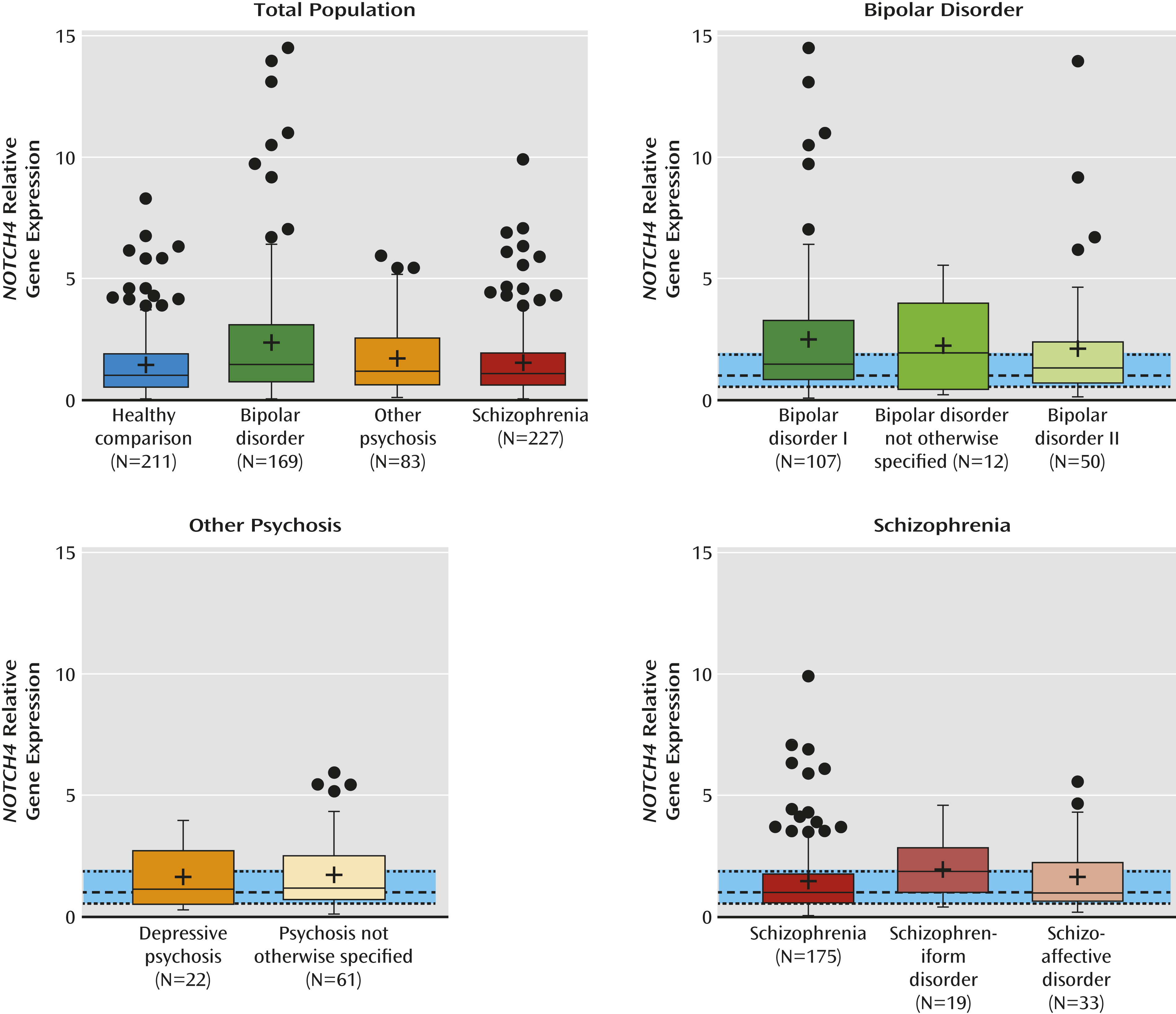
Figure 1,
NOTCH4 mRNA levels were significantly higher in patients with bipolar disorder (mean=2.5, SD=3.3) relative to healthy comparison subjects (mean=1.5, SD=1.4) and patients with schizophrenia (mean=1.6, SD=1.4). Tukey’s post hoc test confirmed bipolar disorder > healthy comparison subjects (p<0.0001) and bipolar disorder > schizophrenia (p=0.001). The difference in
NOTCH4 mRNA levels between bipolar disorder (mean=2.5, SD=3.3) and other psychosis (mean=1.7, SD=1.4) was just above the significance level (p=0.09).
NOTCH4 Expression Adjusted for Confounders
With the exception of lamotrigine, we found no significant effect of medication or number of episodes, hospital admissions, and symptoms on
NOTCH4 expression levels (Table ST1 and Table ST2 in the online
data supplement). Age (t=−3.5, df=689, p=0.001), BMI (t=−3.6, df=629, p<0.001), and C-reactive protein (t=−2.3, df=649, p=0.03) were inversely associated with
NOTCH4 expression, whereas hemoglobin (t=2.1, df=639, p=0.03), lamotrigine (t=2.86, df=689, p=0.004), and alcohol (t=2.8, df=689, p=0.006) were positively associated with
NOTCH4 expression. After entering these as covariates in an ANCOVA model and correcting for multiple testing, only bipolar disorder diagnosis remained significantly associated with
NOTCH4 expression. As summarized in
Table 2, having bipolar disorder explained most of the variance in
NOTCH4 expression after adjusting for a range of confounders.
Increased Expression of NOTCH4 in T Cells From Patients With Bipolar Disorder
In whole blood, erythrocytes and platelets could potentially have contributed to NOTCH4 expression in addition to its expression in leukocytes. However, it is unlikely that the positive correlation between NOTCH4 and hemoglobin reflects an important contribution of erythrocytes to NOTCH4 levels. In fact, the level of erythrocytes was significantly lower in bipolar disorder relative to the other groups (F=9.8, df=3, 640, p<0.01), and we found no detectable NOTCH4 transcript in isolated erythrocytes. In contrast, platelets contained mRNA levels of NOTCH4, potentially reflecting the presence of reticulated platelets. However, we found no significant differences in platelet counts between patients with bipolar disorder (N=146) and comparison subjects (N=203) (bipolar patients: mean=261×109/L [SD=65.10]; comparison subjects: mean=249×109/L [SD=51.64]; p=0.2), and in patients with bipolar disorder, NOTCH4 mRNA levels were negatively and not positively associated with platelet counts (r=−0.15, N=634, p=0.067). Another possibility is that the difference in NOTCH4 mRNA levels between bipolar disorder and the other groups could reflect the difference in leukocyte counts and leukocyte subpopulations. However, we found no significant differences in leukocyte counts between patients with bipolar disorder (N=147) and healthy comparison subjects (N=205) (bipolar patients: mean=6.25×109/L [SD=2.11]; comparison subjects: mean=5.92×109/L [SD=1.63]; p=0.3). Also, in patients with bipolar disorder, NOTCH4 mRNA levels were negatively rather than positively associated with total leukocyte counts (r=−0.22, N=638, p=0.009).
NOTCH4 has been linked to T cell pathophysiology (
15). When analyzing
NOTCH4 expression in isolated T cells and monocytes from 15 patients with bipolar disorder (seven women; mean age=39 years [SD=9]) and 15 sex- and age-matched healthy comparison subjects (seven women; mean age=41 years [SD=11]), we found that T cells contained high
NOTCH4 expression with significantly increased mRNA levels in patients with bipolar disorder (bipolar patients: mean=3.80 [SD=0.90]; comparison subjects: mean=1.00 [SD=0.14] [relative expression normalized for β-actin]; p=0.042). In contrast, monocytes contained only a minor amount of
NOTCH4 mRNA with no differences between patients and comparison subjects (data not shown).
Diurnal Variation and Fasting or Nonfasting Effect on NOTCH4 Expression
Diurnal variation and nonfasting or fasting condition could also have potentially influenced our results. However, the exact times for comparison sample collections were recorded for 188 out of 211 comparison subjects, and we found no significant differences between mean levels of NOTCH4 according to the time when the blood sample was drawn (F=0.68, df=5, 183, p=0.63). Second, the patient collections were all performed within the same narrow time interval (8 a.m.–10 a.m.) and under the same conditions for the three groups of patients. Third, in 15 healthy comparison subjects, blood samples for whole blood extraction were collected at 9 a.m., 1 p.m., and 4 p.m. Although some diurnal variation occurred, with the highest NOTCH4 mRNA levels in the morning, the differences were not statistically significant (effect of time from repeated measures ANOVA, p=0.62). Finally, when testing the influence of fasting by comparing fasting and nonfasting conditions in 15 healthy comparison subjects (9 p.m.), only minor nonsignificant variations were seen (nonfasting: mean=1.00 [SD=1.05]; fasting: mean=0.84 [SD=0.83]; p=0.64).
Association Between Expression Quantitative Trait Loci and NOTCH4 Expression
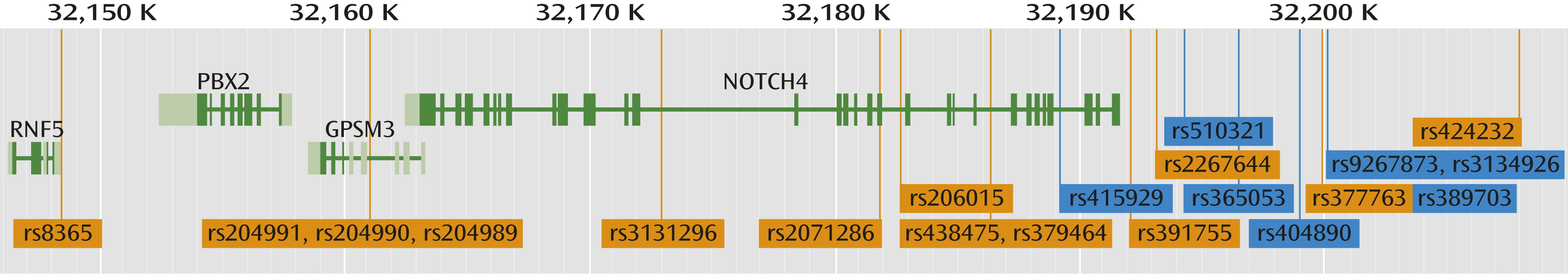
The location of the SNPs investigated is depicted in
Figure 2. Seven SNPs located upstream of the
NOTCH4 gene were associated with
NOTCH4 mRNA expression after using the Bonferroni correction, four of which were in linkage disequilibrium (
Table 3): rs365053, rs404890, rs389703, and rs9267873 (see Figure SF1 in the online
data supplement). We found a significant main effect (Bonferroni-corrected) of SNP on
NOTCH4 expression only in comparison subjects after adjusting for age, BMI, c-reactive protein, hemoglobin, and alcohol (rs510321 [p=1.2×10
−5], rs365053 [p=10×10
−4], rs389703 [p=1.1×10
−3], rs415929 [p=10×10
−3]). In addition, the SNPs rs2071286 and rs3134926 were nominally associated with
NOTCH4 expression in comparison subjects. For most of the SNPs, the allelic effect went in the same direction for all groups (Figure SF2 in the online
data supplement illustrates the direction of allelic effect). We did not find a significant association or a significant interaction effect between SNPs and bipolar disorder (data not shown). This is probably due to inadequate statistical power, because adequately powered GWAS do find such an association (
10).
Discussion
To our knowledge, this is the first study to show higher mRNA levels of
NOTCH4 in whole blood from patients with bipolar disorder relative to healthy comparison subjects, patients with schizophrenia, or patients with other psychotic disorders. In addition, seven SNPs within a 10-kb region upstream of the
NOTCH4 gene were significantly associated with
NOTCH4 mRNA levels. These associations remained significant after controlling for a range of possible confounders and correcting for multiple testing. Our data indicate that the association between
NOTCH4 and bipolar disorder from large GWAS is not restricted to DNA markers, but it is also seen at the mRNA level, supporting the notion that
NOTCH4 is of functional importance in disease pathology. Finally, in line with previous studies on Notch biology in immune cells (
14,
15), our findings suggest that the enhanced expression of
NOTCH4 in the whole blood of patients with bipolar disorder, at least partly, could reflect increased expression in T cells. However, our leukocyte subset data were from relatively few individuals, and caution is needed when interpreting these results.
We identified expression quantitative trait loci to be related to up-regulated
NOTCH4 expression. Previous GWAS have identified an association between the genetic markers in
NOTCH4 in both bipolar disorder and schizophrenia (
2,
11), but we found no association between schizophrenia and
NOTCH4 expression. The reason for this apparent discrepancy is not yet clear, but the association of
NOTCH4 with bipolar disorder both at the DNA and mRNA levels suggests a stronger link between
NOTCH4 and bipolar disorder than with schizophrenia. Whether these
cis-expression quantitative trait loci directly enhance
NOTCH4 activation or influence
NOTCH4 expression through other mechanisms not controlled for in this analysis merits further study. However, our large and well-described sample allowed us to perform an extensive search for confounding factors that could potentially explain why increased
NOTCH4 expression is mainly seen in bipolar disorder and not in schizophrenia. Lamotrigine and high alcohol consumption explained some of the variance. Lamotrigine is a prescribed medication for bipolar disorder, and alcohol use is often associated with bipolar disorder (
20). However, the association between
NOTCH4 mRNA levels and bipolar disorder was seen even after adjusting for these and several other confounders.
NOTCH4 is involved in immune responses, and abnormalities in the immune system have been frequently described in mood disorders. Circulating C-reactive protein may reflect activation of the immune system, and surprisingly, C-reactive protein levels were inversely associated with
NOTCH4 mRNA levels. However, while C-reactive protein is a reliable marker of systemic inflammation, it by no means reflects all upstream inflammatory pathways. In fact, the relationship between
NOTCH4 and the endothelium may not necessarily be reflected by C-reactive protein. We found some indications that
NOTCH4 and C-reactive protein may differently affect vascular endothelial growth factor activity in endothelial cells (
21–
23), potentially suggesting that they may partly reflect different or even opposite pathways. We and others have reported elevated plasma levels of markers of immune activation and signs of increased T cell activation in schizophrenia and bipolar disorder (
6,
7,
9,
10). Mice with a constitutively active
NOTCH4 are characterized by an increase in immature T cells in their bone marrow, with a reduction in B cells. Importantly, there is some evidence that T cell activation itself may influence mood symptoms, as elevated levels of interleukin-2, a T cell product that also activates these cells, is associated with mania (
24). Our finding of enhanced
NOTCH4 expression in T cells in a subset of patients with bipolar disorder may further support a link between enhanced
NOTCH4, T cell activation, and bipolar disorder.
Genes that are subject to regulation by ischemia-hypoxia may play a role in the neurodevelopmental changes observed in the early stages of schizophrenia (
25). The same abnormalities are observed in bipolar disorder at later stages in the course of the illness (
25). In this context, NOTCH signaling pathways play a critical role in vascular development and homeostasis (
26,
27). Results from animal studies have shown that activation of the
NOTCH4 signaling system in the endothelium during embryogenesis causes neuronal cell death and hemorrhage in the cerebral cortex and cerebellum (
17). Furthermore, constitutive
NOTCH4 activation in endothelial cells inhibits angiogenesis, and up-regulation of the NOTCH ligand Delta-like 4 inhibits vascular endothelial growth factor-induced endothelial cell function (
23). This might compromise the vascularization in the adult CNS (
28). Enhanced
NOTCH4 expression could also contribute to endothelial cell activation through the interaction between activated T cells and the endothelium. Indeed, we have seen enhanced levels of von Willebrand factor in bipolar disorder, demonstrating the presence of endothelium-related inflammation in these patients (
10).
NOTCH4 could also be involved in the development of bipolar disorder through more specific neuronal mechanisms. Investigating mRNA levels in postmortem brain tissue is an important and fast-growing field in psychiatric research, but few researchers have specifically investigated
NOTCH4 in bipolar disorder. Using the Stanley Genomics Database (
http://stanleygenomics.org), we found little variation in the expression levels of
NOTCH4 and related genes in bipolar disorder patients (N=49) relative to healthy comparison subjects (N=50) (
29). One microarray study of a Stanley Foundation Brain Collection subsample found indications of
NOTCH4 down-regulation in the frontal cortices with possible association to psychotropic medication (
30), but little is known about the functional mechanisms regulating NOTCH signaling in the brain. One potential mechanism might be the close interaction between the Wnt and NOTCH signaling system during both embryogenesis and adult hippocampal neurogenesis (
31–
33). This is supported by a reported association between bipolar disorder and a gene regulating the Wnt signaling pathway (
34), and genome-wide association results from bipolar disorder were recently found to be related to gene networks involving Wnt and NOTCH signaling (
35). However, additional microarray studies covering more brain regions are needed to determine the role of
NOTCH4 in bipolar disorder neuropathology. Still, the current evidence is in line with the hypothesis that
NOTCH4 has a role in abnormal neurodevelopment that leads to bipolar disorder as well as in the initiation of affective episodes during disease progression.
Some limitations in our study should be addressed. First, this was a cross-sectional study and thus offers no certain explanation concerning causality. On the other hand, our sample is large and very well described, which allowed us to control for a range of confounders. Second, despite clear expression in the brains of healthy comparison subjects, to the best of our knowledge, there is limited information concerning NOTCH4 expression in postmortem brains of individuals with schizophrenia and bipolar disorder, and the quality of gene expression profiling is limited by the lack of live tissue biopsies from the affected area in question. Future research should examine if the elevated NOTCH4 mRNA levels in bipolar disorder could reflect a uniform increase in other relevant tissues, such as the brain. Third, although our primary aim was to determine NOTCH4 mRNA expression levels, and the expression quantitative trait loci analysis was a secondary aim, data on SNP genotyping are missing for some of the individuals who were included in the expression analyses. Fourth, the functional mechanisms of the NOTCH4 SNPs or the splicing variants are not yet known and should be explored in future experimental studies.
The development of
NOTCH4-targeted inhibition therapies has received much attention, particularly in cancer treatment (
36). Our results indicate that patients with bipolar disorder show enhanced
NOTCH4 activation, which provides us with new study opportunities for treatment options in this patient group. In light of evidence indicating bipolar disorder as a neuroprogressive disease (
37), future research should focus on longitudinal studies of the regulatory mechanisms of
NOTCH4 levels in relation to variations in affective symptom levels. Finally, our results indicate that
NOTCH4 expression in postmortem brains of individuals with schizophrenia and bipolar disorder should be further investigated.