Alcoholism is a heterogeneous, complex disorder with multiple subtypes or endophenotypes (
1). Perhaps because of this heterogeneity, the therapeutic effect sizes of the approved medications for the treatment of alcohol dependence have been relatively small (
2). A personalized approach based on the patient’s genetic makeup is increasingly being investigated for delivering optimum treatment to suitable patients. Previously, in a randomized double-blind clinical trial of alcohol-dependent individuals of European descent, we showed that treatment of severe drinking with ondansetron, a specific serotonin-3 (5-HT
3) antagonist, was significantly more efficacious in a subpopulation carrying the serotonin-transporter-linked polymorphic region LL (5′-HTTLPR-LL) and rs1042173-TT (
SLC6A4-LL/TT) genotype combination in the serotonin transporter (5-HTT) gene,
SLC6A4 (
3). Briefly, in that study, we found that
SLC6A4-LL/TT carriers treated with ondansetron had 2.63 fewer standard drinks per drinking day and a percentage of days abstinent 16.99% higher than noncarriers of the
SLC6A4-LL/TT genotypes. In contrast, when patients were not subgrouped according to their 5′-HTTLPR and rs1042173 genotypes, there was no statistically significant difference between the ondansetron and placebo groups.
Although the
SLC6A4-LL/TT genotypes predicted ondansetron treatment response, ondansetron cannot bind directly to 5-HTT molecules because it is an antagonist at the 5-HT
3A receptor subunit (
4–
7). Determination of the 5-HTT genotypes that we selected as being predictive of ondansetron treatment response in our previous study (
3) was therefore indirect and based on previous in vitro and in vivo experimental evidence indicating that
SLC6A4-LL/TT is associated with markedly reduced 5-HTT expression levels in alcohol-dependent individuals (
8–
11). The postsynaptic 5-HT
3 receptors are formed by 5-HT
3A homopentamers or 5-HT
3A and 5-HT
3B heteropentamer (5-HT
3AB) complexes that evince even faster ion transport and conduction. The 5-HT
3B subunits do not bind with serotonin or ondansetron but are important for stabilizing the receptor complex at the cell surface (
12,
13). The 5-HT
3 A and B subunits are encoded by the
HTR3A and
HTR3B genes located adjacently on chromosome 11, separated only by an intergenic region of 28 kb. Hodge et al. (
14), using a transgenic mouse model lacking the gene for the 5-HT
3A subunit, showed that the reduction of alcohol consumption produced by 5-HT
3 antagonism is dependent on the presence of 5-HT
3A-containing receptors. Recently, a genetic association study conducted by Enoch et al. (
15) that examined African American men with a lifetime diagnosis of alcohol, cocaine, or heroin dependence showed an association between these disorders and the particular genotypes of
HTR3A,
HTR3B, and the 5′-HTTLPR. These findings were supported by earlier work from the same group that showed an association between certain
HTR3B genotypes and alcohol dependence with antisocial behaviors (
16).
Discussion
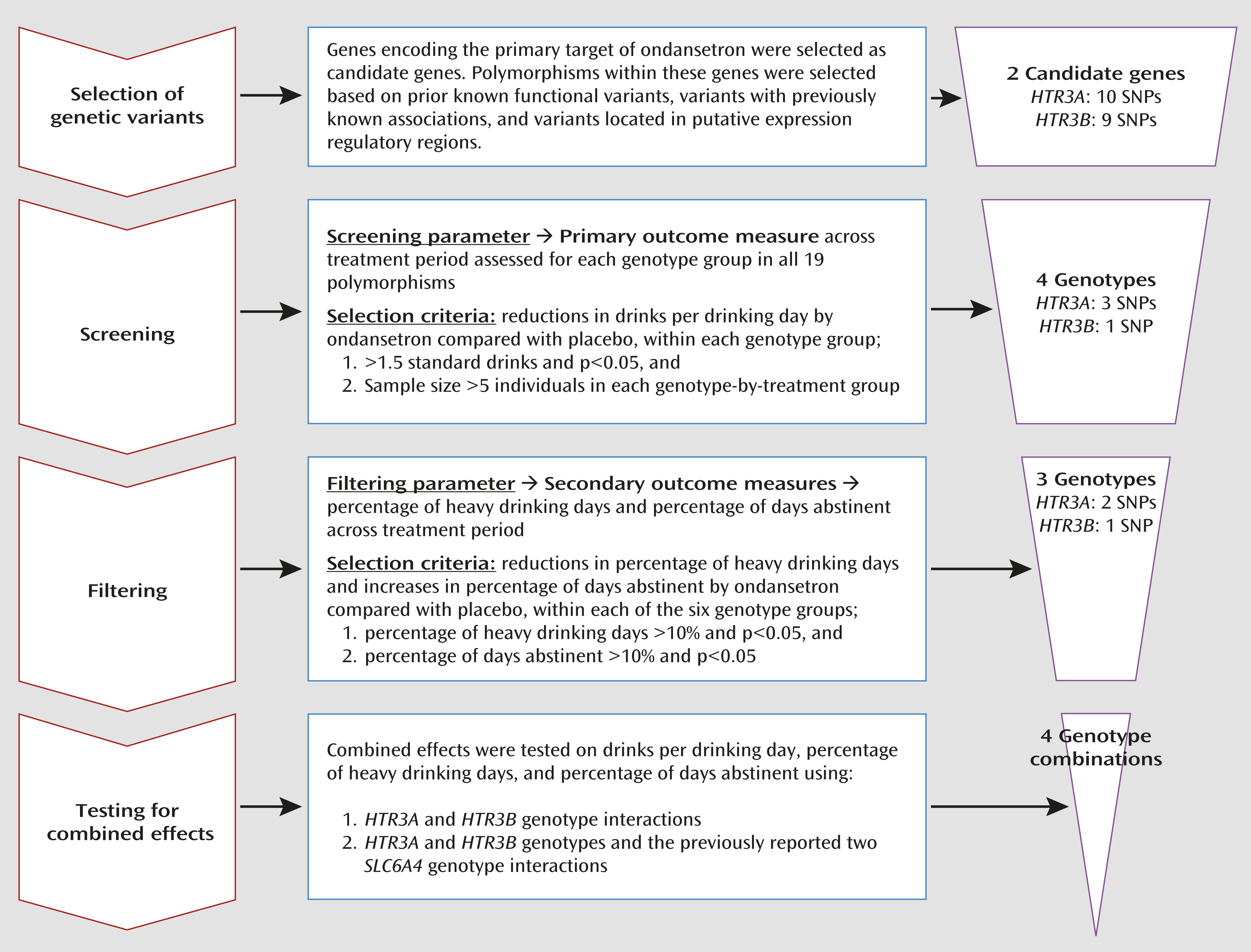
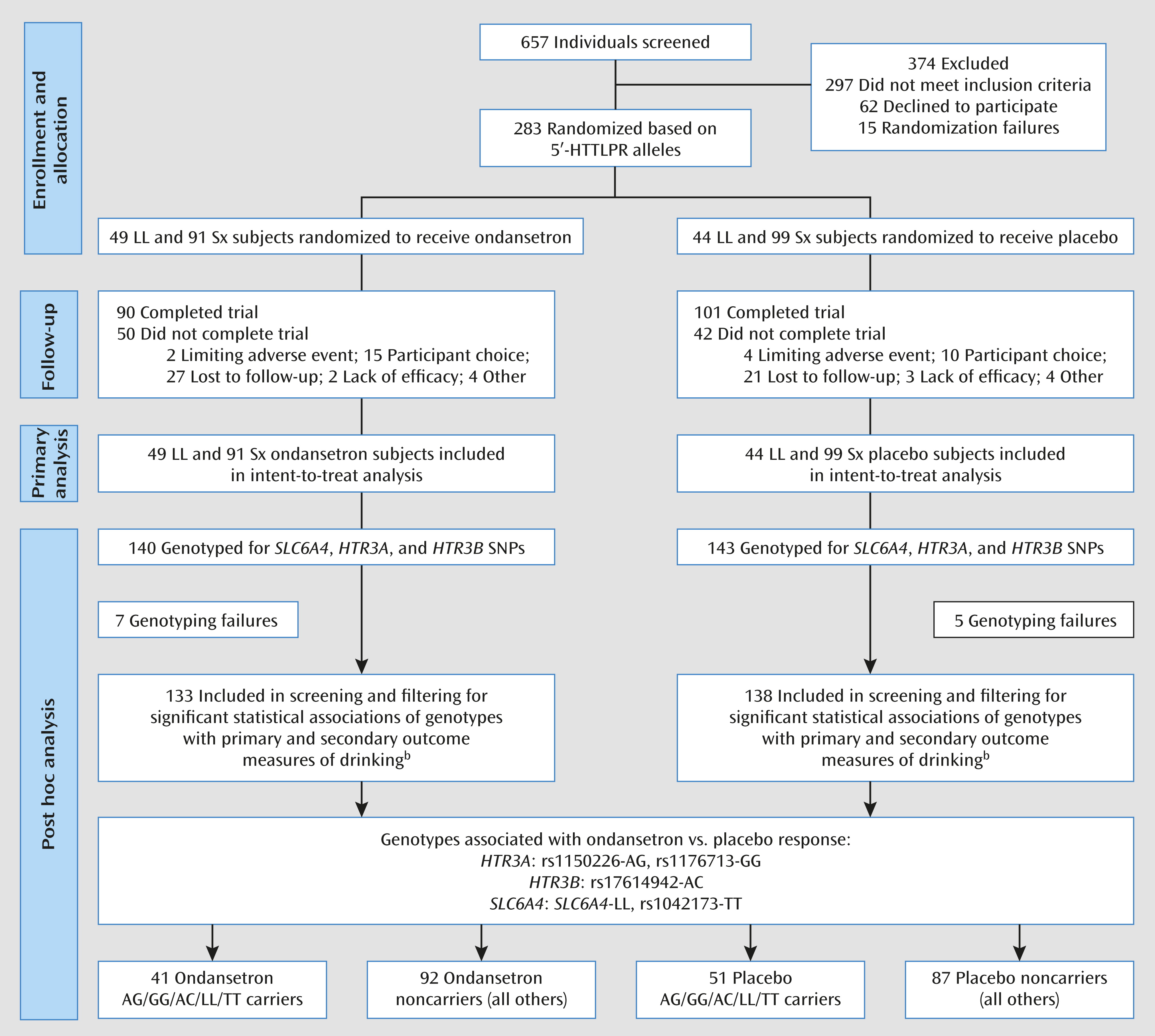
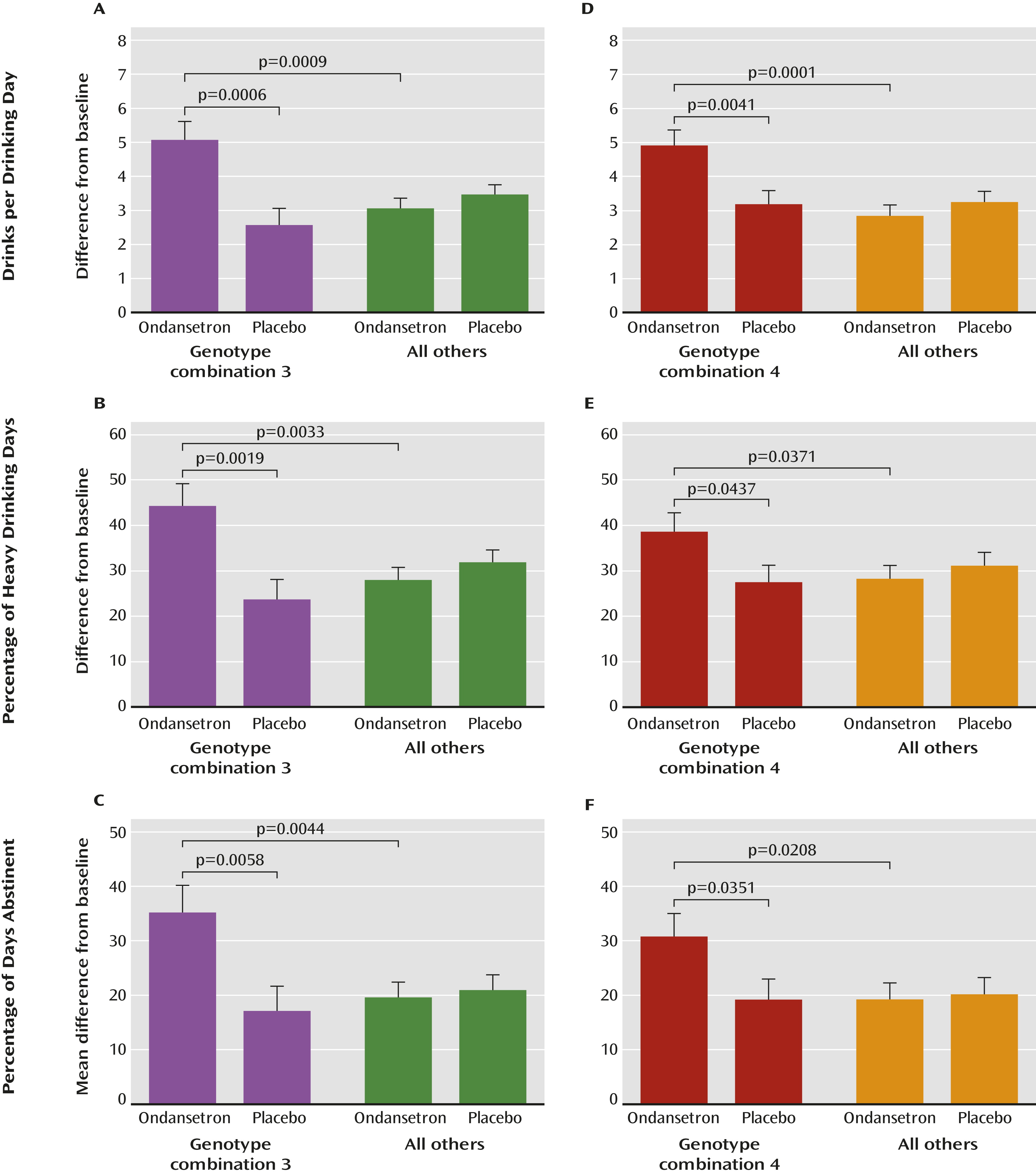
We identified three genotypes in the HTR3A and HTR3B (collectively designated as HTR3) genes that were significantly associated with efficacy of ondansetron treatment for alcohol dependence in individuals with European ancestry. We showed that possession of at least one of the HTR3A-rs1150226-AG, HTR3A-rs1176713-GG, and HTR3B-rs17614942-AC genotypes, along with the previously identified SLC6A4-LL/TT genotypes (i.e., combination 4), was predictive of ondansetron treatment response; approximately 34% of the cohort carried these genotypes.
A unique strength of our findings was that the five-genotype combination 4 (
Table 2) was associated not only with a reduction in the amount of severe drinking (drinks per drinking day) but also with two other measures of treatment response—the frequency of heavy drinking (percentage of heavy drinking days) and of abstinence (percentage of days abstinent). Additional to these widely used outcome measures, we also performed an exploratory analysis for the genotype associations with an extreme dichotomous responder versus nonresponder endpoint efficacy variable, namely, percentage of subjects with no heavy drinking days, which may be used by regulatory agencies as an important outcome measure for larger phase 3 trials (
34). When only one heavy drinking day during the final 4 weeks was allowed, we detected at least a twofold increase in percentage of subjects with no heavy drinking days among participants in the ondansetron group carrying one or more of the combination 4 genotypes compared with participants in the placebo group or noncarriers in the ondansetron group. Notably, however, the carriers of combination 3 who constituted a subset of combination 4 (approximately 25% of the cohort) had more than a fivefold increase in percentage of subjects with no heavy drinking days when the ondansetron and placebo groups were compared, and they might define a group of “super-responders.”
Furthermore, the greatest reductions in drinks per drinking day were seen in small subpopulations possessing both
SLC6A4-LL/TT genotypes and any one or more of the HTR3 genotypes. This finding supports our hypothesis that the combined effects of genetic variants for the presynaptic 5-HTT and postsynaptic 5-HT
3 would predict a greater response than genetic variations within each individual gene, as they represent the state of both synaptic availability of serotonin and its postsynaptic receptor levels. Another factor supporting this hypothesis is the greater effect of
SLC6A4 plus HTR3 than the combined effect of the
HTR3A and
HTR3B genotypes. However, results for the combined effect of
SLC6A4 and HTR3 should be interpreted cautiously, as they were limited by the small sample size within each placebo and ondansetron treatment-by-genotype group (see Table S3 in the online
data supplement), thereby reducing the statistical power to draw definitive conclusions. Nevertheless, these initial statistical trends generally supporting a molecular basis for the use of ondansetron as a treatment for a particular cohort of alcohol-dependent individuals warrant further corroboration using studies with larger sample sizes.
The second-best response for drinks per drinking day was seen in the pooled group of individuals carrying any one or a combination of the three HTR3 genotypes, regardless of LL/TT carrier status (combination 3 in
Table 2). From a biological perspective, this is quite explainable, since 5-HT
3AB receptors are the primary target of ondansetron, which in fact led us to select
HTR3A and
HTR3B as candidate genes for the present study. Even though ondansetron’s therapeutic effect on the selected
SLC6A4 genotypes may appear to be smaller than that on the target HTR3 genotypes, presumably because the biological effect of the 5-HTT on ondansetron’s pharmacodynamics is an indirect one, their interaction is most intriguing in showing a more robust response across different outcome measures of alcohol consumption.
Notably, the significant associations that we detected with SNPs rs1150226 and rs17614942 were through their heterozygous genotypes; that is, the heterozygotes showed significantly greater response to ondansetron compared with both homozygous genotype carriers. Although it is argued that such a comparison utilizing the heterozygous genetic model may not reflect underlying biological mechanism(s), a dominant heterozygous effect over both homozygotes, which is also referred to as molecular heterosis, is an increasingly recognized phenomenon in many human gene-disease association studies that include addictions (
35–
39). Another possible explanation for detecting heterozygous associations could be the small sample sizes of the homozygous minor allele genotype groups, rather than the effects of molecular heterosis.
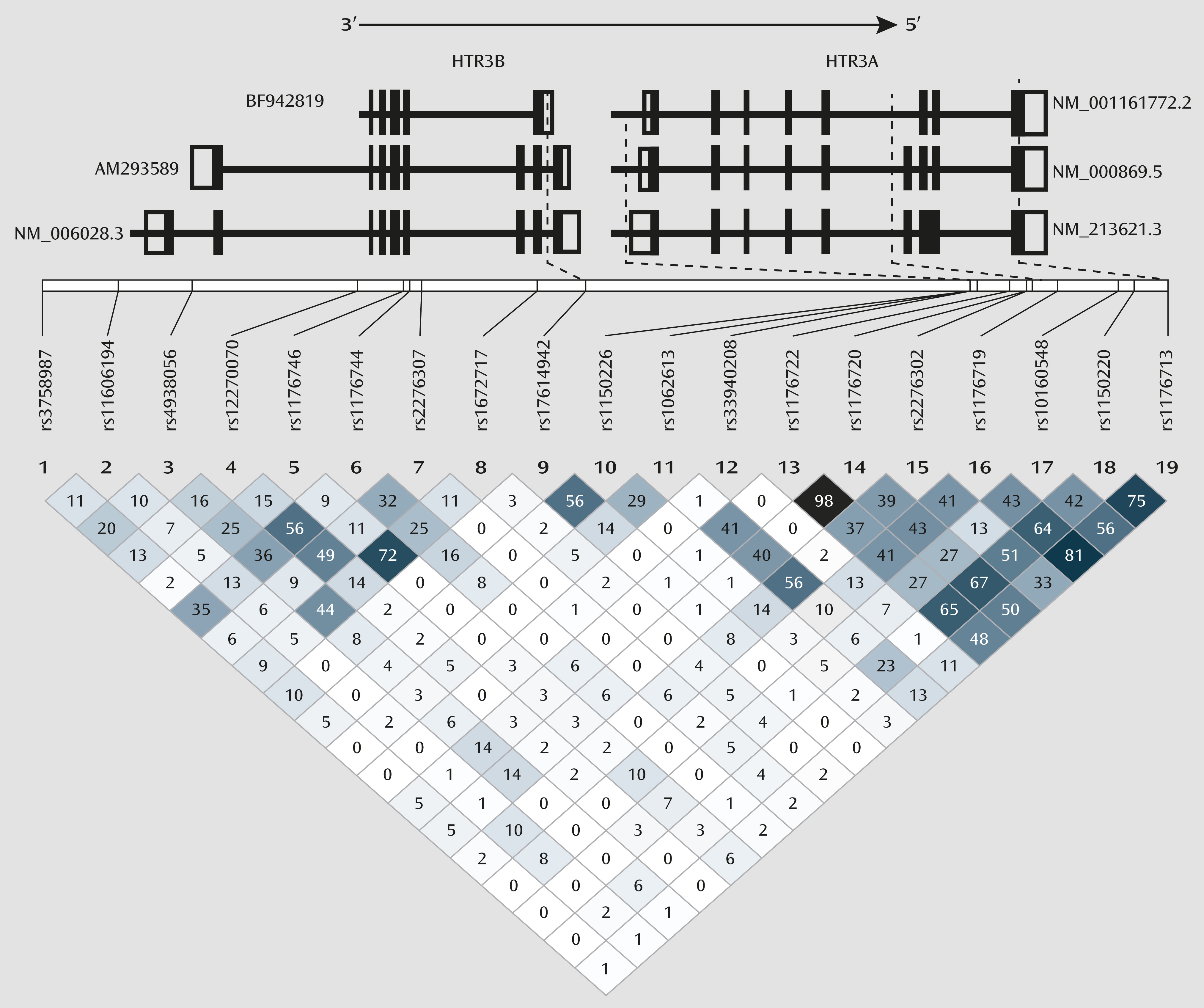
An additional caveat is that the preclinical studies of the functionality of rs1150226 and rs1176713 of
HTR3A and rs17614942 of
HTR3B have not been reported in the literature, and therefore the exact molecular mechanism for their interactive effects remains to be determined. However, given their location within the genes, it is possible that all three of these polymorphisms may alter mRNA expression levels. The rs1150226 is located in the promoter region of
HTR3A within 1 kb of putative binding sites for several transcription factors that have been shown to be modulated by ethanol (
40,
41). Furthermore, genetic association analyses have shown an association of rs1150226 with alcoholism comorbid with antisocial personality disorder (
16), altered temporal lobe activity (
42), and clozapine treatment outcome for schizophrenia (
43). Both rs1176713, which is a synonymous SNP located on exon 8 (Leu459Leu) of
HTR3A, and rs17614942, located in intron 8 of
HTR3B, could alter expression levels of functional 5-HT
3AB subunits through altered splicing of primary mRNA to form nonfunctional secondary mRNA. Interestingly, rs17614942 is located in different regions in different mRNA isoforms (
Figure 3). In primary mRNA isoforms AM293589 and NM.006028.3 (
http://www.ncbi.nlm.nih.gov/IEB/Research/Acembly/av.cgi?c=geneid&org=9606&l=9177), the two longer
HTR3B mRNA transcripts that are found to be present in the brain, rs17614942 is located in intron 8. In rodents, intron 8 consists of a splice acceptor site that results in the long isoform (
44); even though this site is not known to be present in humans, it underlines the importance of the intron 8 region in the formation of various mRNA splice variants. In the shorter primary mRNA isoform BF942819 (
http://www.ncbi.nlm.nih.gov/IEB/Research/Acembly/av.cgi?c=geneid&org=9606&l=9177), which has not yet been reported to be present in the brain, rs17614942 is located in the 3′-UTR, which can also lead to changes in functional 5-HT
3B subunit expression level through different molecular mechanisms, such as altered mRNA polyadenylation, miRNA binding, and decay. In vivo experiments would be required to characterize accurately the functional effects of the variants in combination 4, both individually and collectively. We therefore propose our theorization as a framework on which to launch further molecular investigations (
3).
Another caveat regarding our findings is that the sample size was relatively small for performing statistical comparisons for all different genotype combinations constituted by the five variants included in combination 4. It is therefore not possible to dissect fully the relative contribution of each individual genotype with treatment response. However, an analysis that tested for all 16 different combinations of the five variants was well beyond the scope of this phase 2 clinical trial and would require a multicenter clinical trial that enrolled several thousand treatment-seeking alcohol-dependent individuals.
Another consideration was that we used a descriptive filtering process to control for type I error, by minimizing the number of inferential contrasts, to understand the relationship between ondansetron treatment response and genotype. Thus, an additional replication study is needed in a much larger sample to validate our results. Furthermore, because the frequency of the identified alleles may vary in different ethnic populations, additional studies are needed to test the applicability of our findings to other ethnic populations. Nevertheless, our findings were bolstered by the fact that the combination 4 variants also were included in the detected significant genetic epistatic models by GMDR independent epistatic analyses, which were performed to validate the findings from the primary analysis.
Interestingly, a previous association study conducted by Enoch et al. (
15) suggested that rs1176744 polymorphisms in
HTR3B and
SLC6A4 (L
AL
A) were associated with the development of alcohol dependence and, speculatively, ondansetron treatment response. Confirming these findings, an independent association analysis conducted by our team (
45) also showed a highly significant association between alcohol dependence and a genotype combination comprising
HTR3A-rs10160548,
HTR3B-rs1176744 and -rs3782025,
SLC6A4-rs1042173, and 5′-HTTLPR polymorphisms.
In conclusion, we have shown that polymorphisms in the SLC6A4, HTR3A, and HTR3B genes are predictors of reduced drinking in response to ondansetron. Based on these findings, we hypothesize that the combination 4 genotypes can be used to screen for individuals who would respond to ondansetron. Furthermore, the effect sizes of combination 4 across different measures of alcohol consumption appear to be medium to high. Clinical trials randomized on the basis of combination 4 genotypes are thus needed to test the replicability of our findings.