Neuregulin 1 (NRG1) is a key developmental growth factor that binds to and activates the ErbB class of receptor tyrosine kinases (
1). Differential promoter usage and extensive alternative splicing generate several distinct isoforms of the NRG1 gene, namely types I–VI (
1,
2). NRG1 is a key mediator of multiple neurodevelopmental processes, including cell migration, synaptic formation and plasticity, and myelination (
3). Despite growing evidence demonstrating NRG1’s essential role in the developing murine brain (
4–
7) and its involvement in disorders of neurodevelopment and maturation, including schizophrenia (
8–
11) and bipolar disorder (
12,
13), the developmental expression trajectories of individual NRG1 isoforms during human pre- and postnatal neocortical development are unknown.
Polymorphisms in the NRG1 gene have been associated with risk for schizophrenia in multiple populations. The original risk haplotype (HapICE) was first isolated in the Icelandic population and comprises several single-nucleotide polymorphisms (SNPs), including SNP8NRG243177 (rs6994992) located in the 5′ end of the NRG1 gene; the association was subsequently shown to be relevant to schizophrenia in Scottish, English, Irish, and northern Indian populations (
8–
11). Although NRG1 polymorphisms have yet to be identified in large genome-wide association studies (GWAS) of schizophrenia, likely because of heterogeneity within the gene and across populations (
14,
15), support for association of the NRG1 HapICE region has additionally come from meta-analyses of published data (
16,
17) and three GWAS schizophrenia data sets (
18).
The rs6994992 SNP is located proximal to the 5′ exon (E187), central to a
cis-regulatory element in the NRG1 promoter (
19). Experimental examination of rs6994992, using luciferase promoter assays and site-directed mutagenesis of human DNA constructs, suggests that the SNP is functional (
19). It predicts expression levels of NRG1-IV in the brain, whereby the schizophrenia risk allele (T) is associated with increased levels in the hippocampus and dorsolateral prefrontal cortex (
20,
21). In addition, convergent data from human neurocognitive and imaging studies show that rs6994992 is associated with impairments in neurocognitive functioning and sensory processing, abnormal brain morphology, and reduced neural connectivity (
22–
26). Recently, allelic variation at the locus has been associated with cortical and subcortical neuroanatomical structure in human neonates (
27), indicative of a role for NRG1 in early human brain development. Although the exact neural mechanisms by which the polymorphism is associated with human brain development and functioning remain unclear, previous data implicate a mechanism at the molecular level, involving the NRG1-IV class of neuregulins (
19,
20).
Emerging evidence suggests that variation in genes that are temporally regulated during neurodevelopment and expressed preferentially in the fetal brain are related to the genetic architecture of schizophrenia (
19,
27–
30). Unlike other NRG1 isoforms, NRG1-IV is expressed exclusively in the brain in humans, and its expression is threefold higher in early fetal life (
19). NRG1-IV has been confirmed as a bioactive transmembrane proprotein that is susceptible to proteolytic processing (
31) and regulated by neuronal activity (
4). Novel transcripts of NRG1-IV have been cloned exclusively in human fetal brain tissue. These novel transcripts include fetal variants E and F (
19) (referred to herein as NRG1-IVNV), which share the same E187 leader exon and conserved promoter as NRG1-IV but lack a functional epidermal growth factor and matrix metalloprotease cleavage domain. These findings highlight NRG1-IVNV isoforms as a biologically novel class of early developmental NRG1 proteins, potentially relevant to risk for schizophrenia (
19).
During normal brain development, genes are expressed in patterns specific to developmental stage (
32). Although evidence suggests that total levels of NRG1 mRNA are correlated with postnatal age in the human prefrontal cortex (
33), the expression profiles of the individual major NRG1 isoforms (types I–IV) in human fetal and postnatal neocortical development are unknown. Furthermore, little is known about the transcriptional regulation of fetal-brain-derived splice variants of the NRG1-IV class and whether NRG1-IVNV is expressed in the postnatal human brain and is associated with rs6994992 genotype, as would be predicted given the conserved genetic promoter (
19,
20).
In this study we examined quantitative gene expression profiles of NRG1 types I, II, III, and IV in the prefrontal cortex during human fetal brain development (second trimester) and types I, II, and III across the postnatal lifespan from age 0 to 83 years, and we sought to determine if expression of novel NRG1-IV isoforms, NRG1-IVNVs, is temporally regulated during fetal development, detectable after birth, and associated with genetic variation at the schizophrenia risk polymorphism rs6994992. Subcellular localization and biochemical processing of NRG1-IVNV and NRG1-IV isoforms were examined by using recombinant DNA technologies and heterologous expression systems.
Our results provide novel evidence that NRG1 isoform expression is temporally regulated in the human prefrontal cortex during the second trimester of development and postnatally throughout the lifespan. Specifically, NRG1-I expression was most abundant during the first 2 weeks of the second trimester and declined with fetal gestational age, becoming stable at birth, while NRG1-III increased significantly with fetal gestational age and decreased dramatically after birth to reach stable levels in early adolescence. Additionally, we demonstrate that NRG1-IVNV isoforms are uniquely regulated at the transcriptional level during prenatal and early postnatal development, being expressed after gestational age 15 weeks, through birth, and until 3 years of age, after which they are transcriptionally undetectable. Moreover, expression levels of NRG1-IVNV are associated with rs6994992 genotype, identifying a potential molecular mechanism of early developmental risk for schizophrenia at the NRG1 locus. Finally, we show preliminary qualitative data indicating that NRG1-IVNVs, unlike full-length NRG1-IV (and other transmembrane proprotein NRG1s), are resistant to classic proteolytic processing of the extracellular domain and are enriched within the nucleus of both neurons and human embryonic cells, suggesting that NRG1-IVNV splice isoforms are biologically distinct NRG1-IV proteins that may be key regulators of early cortical development relevant to the pathophysiology of schizophrenia.
Method
Human Postmortem Tissue
Postmortem human brains from the National Institute of Mental Health (NIMH) Clinical Brain Disorders Branch were obtained at autopsy from the Washington, D.C., and northern Virginia medical examiners' offices, all with informed consent from the legal next of kin (protocol 90-M-0142 approved by the NIMH/NIH institutional review board). Additional postmortem fetal, infant, child, and adolescent brain tissue samples were provided by the National Institute of Child Health and Human Development’s Brain and Tissue Bank for Developmental Disorders (
www.BTBank.org) under contracts NO1-HD-4–3368 and NO1-HD-4-3383. Samples from 195 normal control individuals who ranged in age from 0 to 83 years were available for this study. Their mean age was 30.73 years; they included 59 females and 136 males; there were 101 African American, 85 Caucasians, five Hispanics, and four Asians; the mean postmortem interval was 30.46 hours (SD=16.54); the mean pH was 6.54 (SD=0.31); and the mean RNA integrity number was 8.3 (SD=0.9). A total of 41 fetal brain samples were available from gestational weeks 14–39; they were from 21 males and 20 females, 39 African American and two Caucasian; the mean postmortem interval was 2.47 hours (SD=2.10); and the mean RNA integrity number was 8.9 (SD=1.1). The institutional review board of the University of Maryland at Baltimore and the State of Maryland approved the protocol, and the tissue was donated to NIMH under the terms of a material transfer agreement. Processing of tissue from both tissue banks was handled in the NIMH laboratory by the same team of investigators, as described previously (
32). Normal control subjects were defined as individuals with no history of psychiatric symptoms, psychiatric diagnosis, psychiatric admission, or substance abuse. All subjects were also without a neuropathological diagnosis and had negative toxicology test results (
34). Further details of brain dissections and sample preparation are described in the supplemental text in the
data supplement accompanying the online version of this article.
Real-Time Quantitative PCR
Quantitative gene expression levels were measured by real-time polymerase chain reaction (PCR) by using an ABI Prism 7900 sequence detection system with 384-well format (Applied Biosystems, Foster City, Calif.) and quantification by means of the standard curve method, as described previously (
20). Specific primer and isoform-specific TaqMan probe combinations were used to assess mRNA expression levels of NRG1 types I–IV in the prefrontal cortex, as previously described (
20). Additional primer and isoform-specific probe sets were designed to amplify NRG1-IVNV isoforms on the basis of the structure of novel fetal variants E and F cloned by Tan et al. (
19) (accessions EF372277 and EF3372275, respectively). Details of the NRG1-IVNV primers and probe design and genetic location are shown in Table S1 and Figure S1 in the online
data supplement. No other known NRG1 transcript lacks the β and stalk coding exons; therefore, the real-time PCR probe designed to span these exons selectively detects NRG1-IVNVs. Expression levels of the NRG1 isoforms were normalized to the geometric mean of the expression of three control genes (β-actin, GAPDH, and PBGD), as described previously (
35) and in the supporting text in the online
data supplement.
Determination of rs6994992 Genotype
The rs6994992 polymorphism was genotyped because of its previous association with schizophrenia and with brain expression of full-length NRG1-IV (
8–
11,
20,
21). Genotyping was performed by using the TaqMan 5′-exonuclease allelic discrimination assay (Life Technologies, Carlsbad, Calif.) (details available on request). Genotype reproducibility was routinely assessed by regenotyping all samples and was generally greater than 99%. The overall genotyping failure rate was less than 1%. DNA genotype was determined from cerebella of fetal and postnatal samples up to 3 years of age, by means of previously described methods (
20) and the following custom-designed SNP assay: forward primer 5′AATTAGTAGGATTGGATGTTTGAACCA 3′, reverse primer 5′ GATGGAGCGCTTCAGGAGAA 3′, probe 1 5′ FAM-CCAGTATACgTTCACTTG-MGB 3′, probe 2 5′ VIC-CCAGTATACaTTCACTTGA-MGB 3′. Genotype analysis was restricted to NRG1-IVNV mRNA. Fetal samples from ages 14 and 15 weeks were excluded from the analysis because of low detectable expression of NRG1-IVNV. Thus, 34 individuals (gestational age, 16 weeks and above) were used for genotypic analysis.
Generation of NRG1-IV Splice-Variant-Specific Recombinant DNA Constructs
Human NRG1-I-β1, full-length NRG1-IV (NRG1-IV-β1a) (Genbank accession EF372273), and NRG1-IVNV (accession EF372275)
N-terminally c-Myc-tagged DNA constructs were generated as described in detail in the supporting text in the
data supplement accompanying the online version of this article. NRG1-I-β1 was generated as a positive control for comparison with full-length NRG1-IV-β1a, given that both isoforms are single-pass NRG1 transmembrane proproteins and have previously been shown to have similar subcellular distributions (
31).
Analysis of Membrane Targeting and Proteolytic Processing of Human NRG1-IV Isoforms in Heterologous Cells
Proteolytic processing of transmembrane neuregulins (i.e., NRG1-I-β1) and subsequent release of the extracellular epidermal growth factor domain is regulated by activation of protein kinase C (PKC) with the phorbol ester phorbol 12-myristate 13-acetate (PMA) (
36,
37). The susceptibility of NRG1-IV variants to stimulated proteolytic processing was studied in human embryonic kidney (HEK293) cells transiently transfected with NRG1-IV-β1a or NRG1-IVNV as described in the supporting text in the online
data supplement.
Subcellular Localization of NRG1-IV and NRG1-IVNV in Rat Primary Neurons and HEK293 Cells
Rat hippocampal neurons and HEK293 cells were transiently transfected with c-Myc-tagged DNA constructs encoding NRG1-I-β1, NRG1-IV-β1a, or NRG1-IVNV and immunostained with an antibody to human c-Myc, as described in the supporting text in the online
data supplement. Qualitative comparison of the subcellular localization of NRG1 variants was performed in neurons and heterologous cells by means of confocal microscopy.
Statistical Analyses
Statistical analyses were performed by using IBM Statistics SPSS, version 21 (IBM, Amonk, N.Y.). The relationships between expression levels of NRG1 types and demographic variables, including age, postmortem interval, pH, and RNA integrity number, were tested by using Spearman’s test of correlation, separately in the fetal and postnatal study cohorts. Effects of sex and race on NRG1 isoform expression were examined with analyses of covariance controlling for age. Effects of genotype at rs6994992 on NRG1-IVNV expression levels were explored by using the nonparametric Kruskal-Wallis test with genotype as an independent factor. To explore potential confounds in the rs6994992 association with NRG1-IVNV expression, post hoc analyses of variance (ANOVAs) were conducted in the three genotypic groups separately to assess the effects of age, sex, and where warranted, race.
Results
Developmental Expression of NRG1 Type I-IV and NRG1-IVNV Isoforms in Fetal Prefrontal Cortex
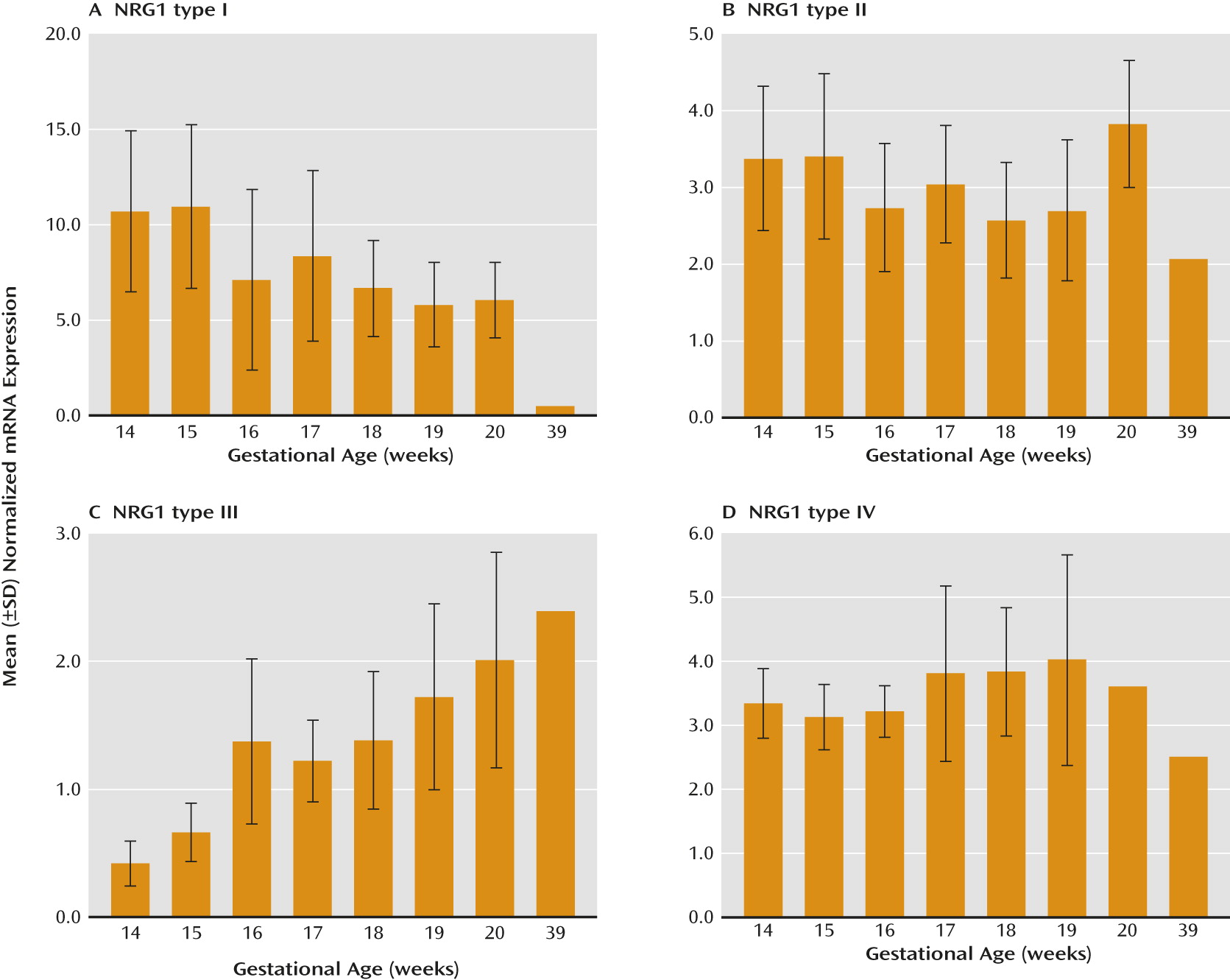
Developmental profiling of transcripts encoding NRG1 isoforms I–IV in the prefrontal cortex during human neocortical development (gestational age, weeks 14–39) revealed that NRG1 types I–IV are tightly regulated and somewhat distinct. NRG1-I mRNA expression was highest at the beginning of the second trimester and subsequently decreased with gestational age (r=−0.49, N=41, p=0.01). In contrast, NRG1-III exhibited an opposite trajectory, being lowest at the beginning of the second trimester and significantly increasing with gestational age (r=0.61, N=41, p≤0.0001) (
Figure 1, parts A and C). Expression of NRG1-II and NRG1-IV showed no significant correlation with gestational age (r=−0.25, N=41, p=0.12, and r=0.06, N=41, p=0.70, respectively) (
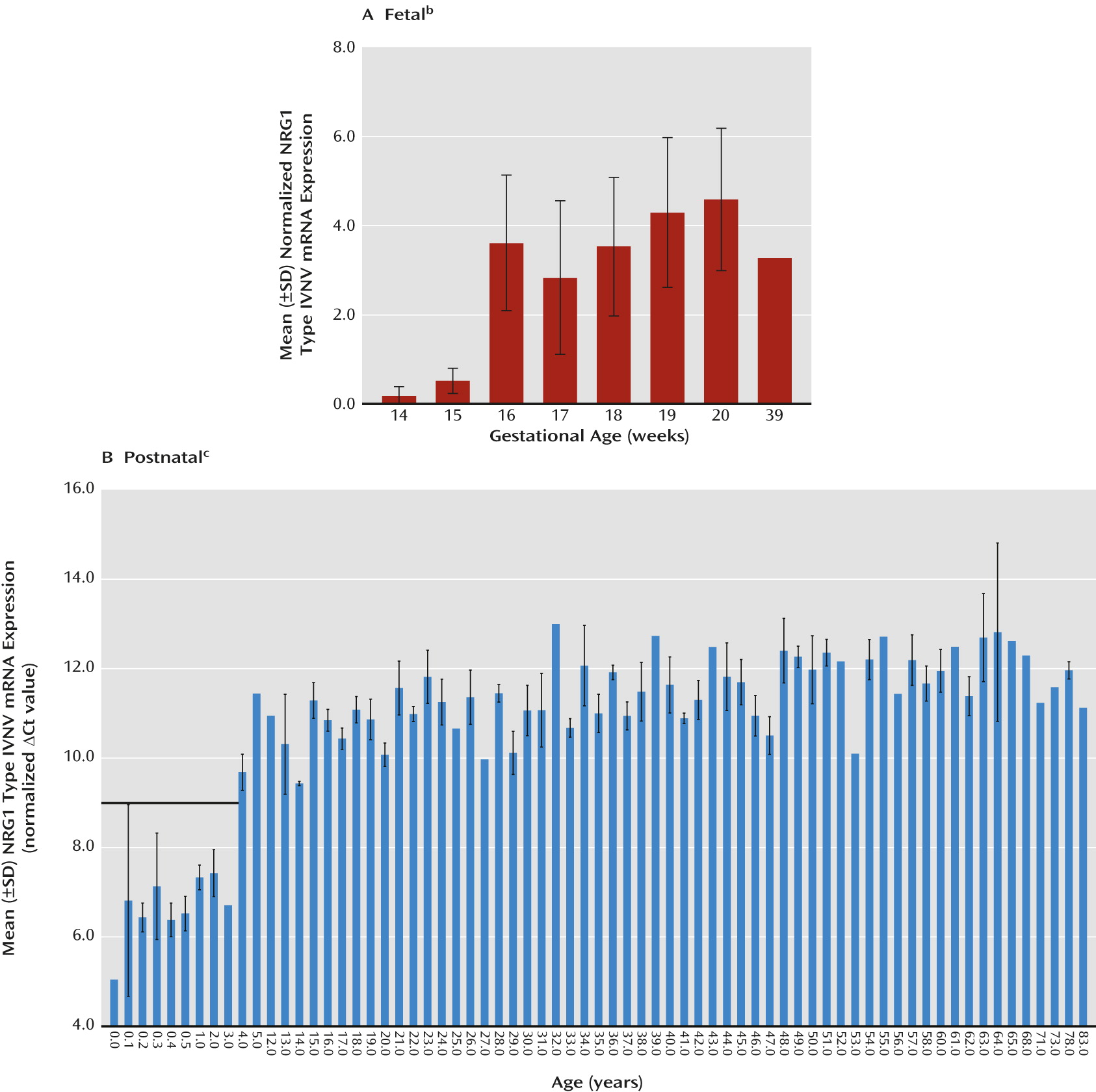
Figure 1, parts B and D). We note that the observed lack of developmentally regulated changes in NRG1-IV expression is potentially confounded in fetal development because the primer and probe used to span the E187 and Ig exons will additionally detect NRG1-IVNV. To wit, targeted amplification of NRG1-IVNV isoform expression during fetal prefrontal cortex development revealed that this novel class of NRG1-IV isoforms increases significantly with age during the second trimester (r=0.63, N=41, p≤0.00001) (
Figure 2A), with the highest levels observed after 15 weeks gestation. No significant correlations were observed between RNA integrity number and NRG1 isoform expression (range: r=0.15 to –0.15, p=0.24–0.92), between postmortem interval and NRG1 isoform expression (range: r=0.05–0.14, p=0.69–0.97), or between RNA integrity number and gestational age (r=−0.13, p=0.93). No effects of sex were observed (p>0.20). The ranges of values for cycle threshold (Ct), the number of cycles at which fluorescence achieves a defined threshold of detection, are reported in the supplementary results in the online
data supplement for each isoform.
Temporal Dynamics of NRG1 Isoform Expression in Prefrontal Cortex Across Postnatal Lifespan
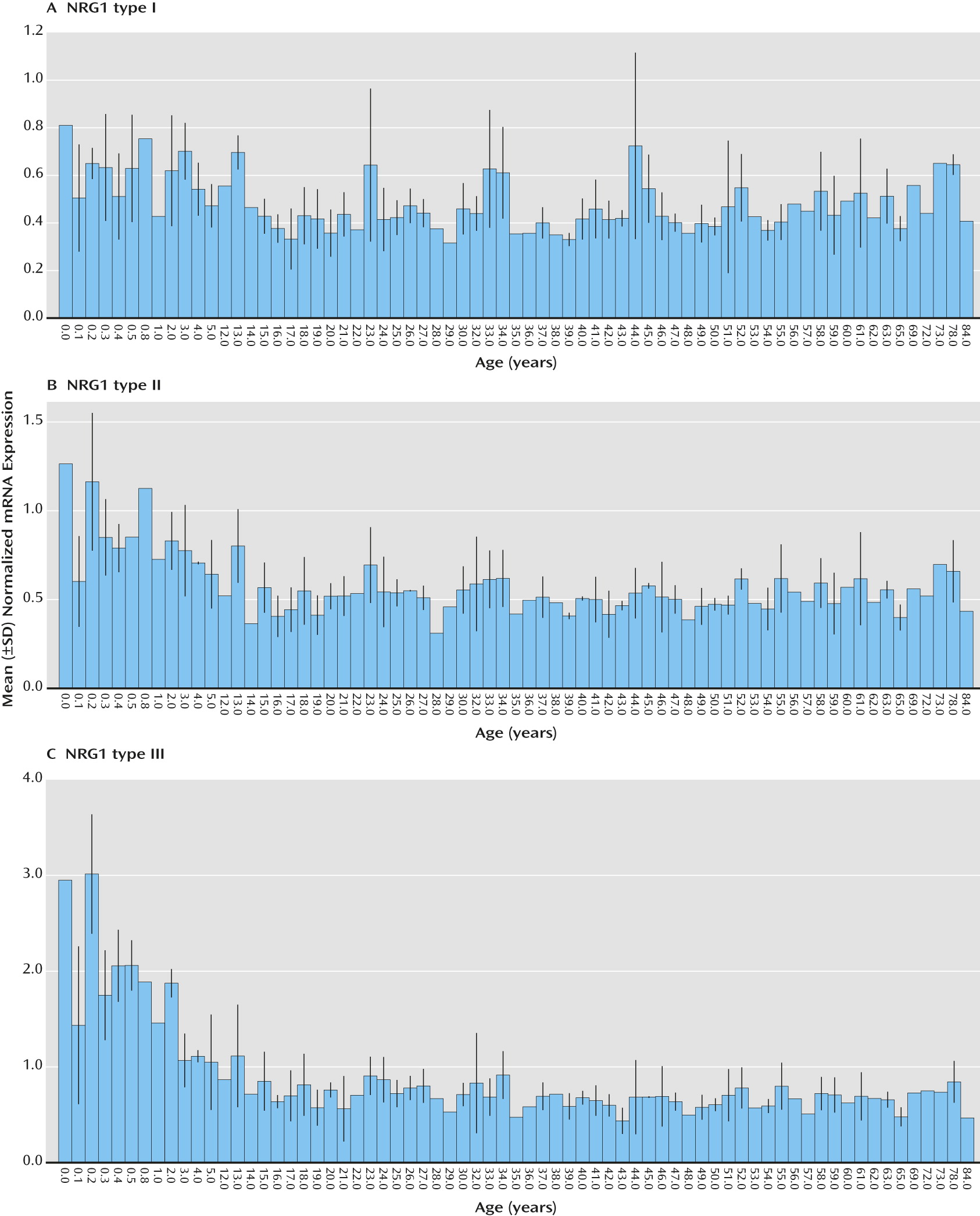
Analysis of quantitative expression trajectories of NRG1-I, -II, and -III across postnatal development revealed significant inverse correlations of age and expression of NRG1-II and NRG1-III, whereby expression was highest at birth and significantly decreased with age (type II: r=−0.26, N=189, p<0.001; type III: r=−0.47, N=189, p<0.000001) (
Figure 3, parts B and C). Expression of NRG1-I, in contrast, was essentially stable across postnatal development and aging, showing no correlation with age (
Figure 3, part A). Previous investigations indicate that NRG1-IVNV isoforms may be exclusively transcribed in the fetal brain, as suggested by the absence of expression in library screens of adult human brains (
19). Remarkably, postnatal expression of NRG1-IVNV was detected at birth and in early infancy and decreased dramatically with age, becoming undetectable after 3 years (cut-off Ct >31; r=0.40, N=25, p=0.01;
Figure 2, part B). There was no significant effect of race or sex on any NRG1 isoform mRNA expression (p>0.20) and no correlation of pH or postmortem interval with individual NRG1 isoform expression (p>0.10). RNA integrity number correlated weakly but significantly with individual NRG1 isoform expression (range: r=0.17–0.23, N=189, p<0.01). However, RNA integrity number did not correlate with age (r=−0.33, p=0.68), and controlling for RNA integrity number in a partial correlational analysis confirmed significant effects of age on type II (r=−0.34, N=189, p<0.001) and type III (r=−0.53, N=189, p<0.000001) NRG1 expression. A secondary post hoc analysis by ANOVA examining the effects of age, controlling for sex, race, pH, postmortem interval, and RNA integrity number, on NRG1 type I–III expression across postnatal development confirmed significant effects of age on NRG1 types II and III (see additional results in the online
data supplement). There were no significant correlations of age with the expression of the individual housekeeping genes (GAPDH, β-actin, and PBGD) or with the geometric mean (r=−0.079–0.099, p>0.18). Ranges of Ct values for each isoform are reported in the online supplementary results.
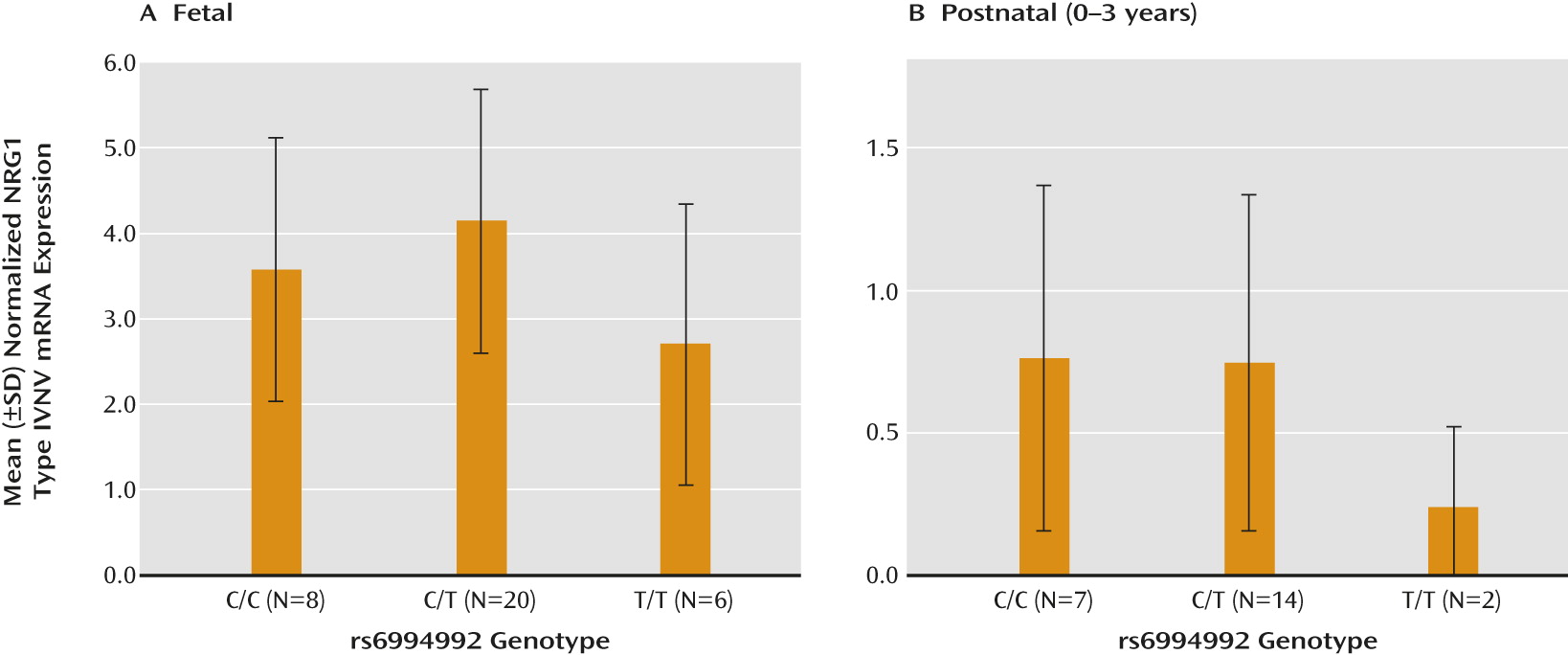
Effect of rs6994992 Genotype on NRG1-IVNV mRNA Expression in Prefrontal Cortex
Given the previous association of rs6994992 with full-length NRG1-IV mRNA expression in the adult human brain (
20,
21), we investigated its association with expression of the novel NRG1-IVNV splice isoform in the developing brain. The effect of rs6994992 genotype on NRG1-IVNV mRNA expression was examined in the prefrontal cortex of fetal samples from gestational age weeks 16–39 (N=34) and postnatal samples ranging from birth to 3 years of age (N=23), when NRG1-IVNV expression was apparent (
Figure 2, parts A and B). The rs6994992 genotype was significantly associated with NRG1-IVNV expression in the fetal prefrontal cortex, and there was a nearly significant association, with the same allelic directionality, in early postnatal life (main effect of genotype: H=7.17, df=2, p=0.03, and H=4.78, df=2, p=0.08, respectively). Individuals homozygous for the risk allele (T) exhibited lower cortical levels of NRG1-IVNV mRNA than did individuals carrying the nonrisk allele (C) (
Figure 4, parts A and B). Post hoc ANOVA exploring the impact of age and sex on NRG1-IVNV expression separately in the three genotypic groups revealed no significant effect of either factor in either the fetal cohort (sex: p>0.12; age: p>0.29) or postnatal cohort (sex: p>0.19; age: p>0.20). No significant effects of race on NRG1-IVNV expression across genotype were observed (p>0.07).
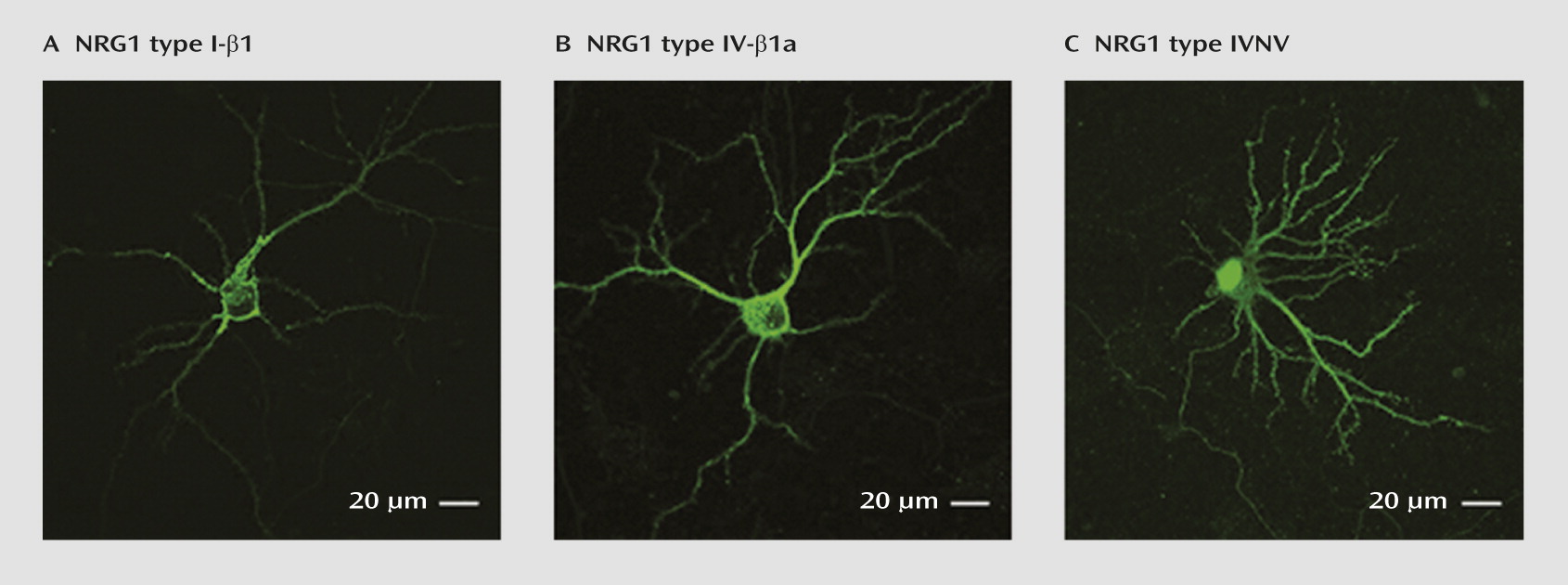
Subcellular Localization of NRG1-IV and NRG1-IVNV Isoforms in Rat Hippocampal Neurons In Vitro
NRG1-IVNV isoforms represent a novel class of NRG1 isoforms that are predicted to give rise to truncated NRG1 proteins that lack a functional transmembrane domain (
19). However, direct experimental evidence for this is lacking. We used confocal microscopy to qualitatively compare the subcellular distribution of 5′ c-Myc-tagged human DNA constructs encoding NRG1-I-β1, NRG1-IV-β1a, and NRG1-IVNV in transiently transfected hippocampal neurons. Immunofluorescence revealed that NRG1-IV-β1a and NRG1-I-β1 proteins were targeted to the cell membrane and distributed in a punctate manner throughout the cell soma and proximal dendrites (
Figure 5, parts A and B). In contrast, NRG1-IVNV was more diffusely present throughout the cell body and dendrites and highly localized to the nucleus (
Figure 5C). Immunofluorescent staining confirmed enrichment of NRG1-IVNV within the cell nucleus of transiently transfected HEK293 cells, as observed by a high degree of colocalization with the nuclear marker DAPI (see Figure S2 in the online
data supplement).
Effect of Proteolytic Processing Mediated by PMA Stimulation on Cell Surface Protein Expression of NRG1-IV and NRG1-IVNV
HEK293 cells transiently expressing NRG1-IV-β1a or NRG-IVNV were established. Western blot analysis of whole cell protein extracts revealed an immunoreactive band of approximately 66 kDa for NRG1-IV-β1a, consistent with the predicted molecular weight of NRG1-IV-β1a (
19) (see Figure S3A, lane 3, in the online
data supplement). Lysates from cells transfected with NRG1-IVNV confirmed translation of truncated protein products of the predicted size (approximately 29 and 31 kDa in fetal variants E and F, respectively, as shown in Figure S3A, lanes 4 and 5, in the online
data supplement), in line with the fact that these NRG1-IV variants contain a premature 3′ stop codon (
19). No immunoreactive bands were present from lysates of untransfected HEK cells or cells transfected with an empty vector (online Figure S3A, lanes 1 and 2). NRG1-IVNV isoforms, unlike the NRG1-IV proprotein, lack a matrix metalloproteinase domain, which is necessary for proteolytic processing of the extracellular domain (
19). To experimentally assess the proteolytic processing susceptibility of NRG1-IV transcripts, we transiently transfected HEK293 cells with either 5′ c-Myc-tagged NRG1-IV-β1a or NRG1-IVNV (type IV fetal variants E and F [
19]), stimulated them with PMA, and collected the conditioned media. PMA stimulation induced release of a 30-kDa extracellular NRG1 fragment in cells transfected with NRG1-IV-β1a (online Figure S3B, lane 6). In contrast, no immunoreactive bands were observed in media collected from PMA-treated cells transfected with NRG1-IVNV (online Figure S3B, lanes 7 and 8). Vehicle-treated cells also showed no immunoreactive band, confirming PMA stimulation as the mechanism of extracellular fragment release (online Figure S3B, lane 5).
Discussion
The trajectory of normal brain development occurs in multiple stages that span the embryonic, neonatal, and early postnatal periods into adolescence and adulthood (
38,
39), with each critical window of development being genetically determined, epigenetically regulated, and environmentally influenced. Research has generated many putative psychiatric risk genes, some of which fall into critical neurodevelopmental pathways (
4–
7,
27–
30), but the transcriptional mapping of the normal developmental expression trajectories of these genes across human brain development and the mechanisms by which risk genetic variants could alter these courses largely remain unclear.
This is the first study, to our knowledge, to describe the individual temporal dynamics of NRG1 isoform (i.e., types I-IV and NRG1-IVNV) expression during human fetal and postnatal neocortical development and reveals potential novel mechanisms of the role of NRG1 in early cortical development. Furthermore, our study characterizes a novel class of NRG1-IV variants, NRG1-IVNV, which in the human brain are developmentally and genetically regulated, expressed exclusively during critical early developmental periods, and represent a biologically distinct subclass of NRG1 proteins, with relevance to risk for schizophrenia.
Although we have refrained from inferring direct comparative abundance of the individual NRG1 isoforms at different stages of development, primarily because even small differences in PCR efficiencies across targets can distort relative expression measurements (
40), we note that the Ct values for NRG1-IVNV in the fetal brain (after 15 weeks) are in the range of 25–27. Values less than 29 represent strong positive reactions indicative of abundant target nucleic acid, suggesting that NRG1-IVNV isoforms represent relatively abundant isoforms of NRG1, with key roles during early neurodevelopment. Further studies are needed to determine the regional and cell-type-specific expression of NRG1-IVNV during human brain development.
Regarding expression of NRG1 isoform types I, II, and III during human brain development, we found that NRG1-I expression peaks during the early second trimester of brain development and decreases with fetal gestational age. NRG1-1 was stable from birth throughout the postnatal lifespan. In contrast, NRG1-II was stable throughout fetal development and decreased after birth, to reach stable levels in early adolescence that continued across aging, whereas NRG1-III increased dramatically during the second trimester, was expressed prominently at birth, and declined dramatically in the periadolescent period, to become stable across the lifespan. Overall, our observations are broadly consistent with expression patterns observed in developing rat cerebral cortex (
4)—whereby NRG1-II and III were shown to be highest in the early neonatal period (P5), declined by P15, and became stable across development—and those seen in mice (
41).
NRG1 has a diverse range of functions during neurodevelopment (
1,
3), and our findings provide further support that NRG1 isoforms, in particular NRG1-I and NRG1-III, play distinct roles in human cortical development and maturation, including neuronal migration, synaptogenesis, gliogenesis, myelination, and the development of glutamatergic, GABA-ergic, and cholinergic neurotransmission (
42,
43). For example, the steep trajectory of increase in NRG1-III expression in the second and third trimester and the decreasing level after adolescence are potentially consistent with the course of myelination, which in humans begins around week 28 and continues into adolescence (
39). It is interesting that the temporal dynamics of NRG1-IVNV expression were similar to those of NRG1-III, with the exception that NRG1-IVNV was undetectable after the first 3 years of life, a postnatal period critical to the elaboration of neuronal processes and synaptogenesis (
39).
The rs6994992 risk genotype predicts elevated expression of full-length NRG1-IV in the adult brain (
20,
21). In this study we explored the genetic association of developmentally regulated NRG1-IV splice isoforms, i.e., NRG1-IVNV, with rs6994992. Unexpectedly, we observed that the schizophrenia-risk genotype is associated with lower expression of NRG1-IVNV during the fetal and early neonatal critical period. The mechanisms behind the differential regulation of NRG1-IV isoform expression as it relates to the rs6994992 locus remain unknown, but they may involve interaction of the
cis effects of rs6994992 with epigenetic regulation of splicing of the NRG1-IV pre-mRNA, a mechanism that plays key roles in regulation of splicing during early development (
44). The dichotomous expression association suggests that a balance between NRG1-IVNV expression and full-length NRG1-IV expression is required throughout neocortical development. This suggestion is consistent with a potential molecular mechanism underlying the association of rs6994992 with cortical and subcortical structure in the human neonate (
27). While initial data suggest that rs6994992 is a functional polymorphism (
19), we also note that it may be in linkage disequilibrium with an unknown causal variant in the NRG1 promoter, which modulates expression of NRG1-IV and NRG1-IVNV and risk for schizophrenia. Finally, in demonstrating a transcriptional association of a schizophrenia susceptibility variant during early fetal brain development, we have identified a potential molecular mechanism of early developmental risk at the rs6994992 NRG1 locus and provide support for schizophrenia as a disorder of neurodevelopment (
45).
Proteolytic cleavage of the extracellular domain of neuregulins allows paracrine activation of ErbB receptors (
1), and this study provides evidence consistent with this for NRG1-IV. Remarkably, NRG1-IVNV isoforms were insensitive to PMA-induced cleavage, as predicted from their structure (
19). The differential localization and resistance to proteolytic cleavage of NRG1-IVNV suggests that these variants have a unique function within the cell compared with NRG1-β1 isoforms. Like NRG1-IVNV, other truncated C-terminal NRG1 isoforms also display resistance to proteolytic processing and are confined to the nucleus and intracellular organelles (
46–
49). It is intriguing that some isoforms of NRG1 have been shown to translocate to the nucleus, where they act as transcriptional regulators, altering the activity and expression of target genes (
50,
51). Given its biochemical structure and nuclear enrichment, we propose that NRG1-IVNV may act as a transcription factor, rather than a bioactive NRG1. In keeping with this hypothesis, alternative splicing of β and stalk exons in NRG1-IVNV transcripts alters the amino acid sequence so that only four of the six cysteine epidermal growth factor residues are present, and thus epidermal growth factor bioactivity is predicted to be lost (
19).
Taken together, our observations provide novel insights into the transcriptional and genetic regulation of NRG1s during normal human brain development, and they identify NRG1-IVNV as a novel, diverse class of developmentally regulated growth factors associated with schizophrenia susceptibility. Future studies are required to determine the neurobiological role of NRG1-IV and NRG1-IVNV and mechanistically how they relate to the development of schizophrenia.
Acknowledgments
The authors thank Dr. Thomas M. Hyde for his efforts in human brain tissue collection, anatomical dissection, and demographic characterization; Dr. Daniel Weinberger for research support; Dr. Wei Tan for construction of the human cDNA constructs; Dr. Tian Zhang Ye for computational support; and Dr. Bhaskar Kholachana for genotyping of rs6994992.