Antipsychotics were incidentally discovered more than 60 years ago, but their mechanisms of action have still not been fully revealed. Antipsychotics are the main medication available to patients with schizophrenia but have little effect on negative and cognitive symptoms of the disease. Current antipsychotics are also limited by serious side effects (
1,
2) that reduce treatment compliance (
3), and about one-third of patients with psychosis are classified as treatment resistant (
4). Thus, there is great need for improved medications for these patients, but drug development has been hampered by poor knowledge of disease etiology and underlying genetics. There is a growing interest in harnessing knowledge of risk genes for developing better treatments for common diseases (
5–
7). For example, Nelson et al. (
8) found that drugs with genetically supported mechanisms of action succeeded in moving from phase 1 trials to gaining approval twice as often as drugs without genetic support. Schizophrenia is highly heritable (
9), and a well-powered genome-wide association study (GWAS) identified as many as 108 independent loci related to the disease, containing more than 300 genes (
10). Improved knowledge of these risk genes may be used to inform drug development by revealing potential mechanisms of action of current drugs and by identifying new drug targets. Antipsychotics bind to numerous proteins (
11), of which dopamine and serotonin receptors are the only ones with known biological links to schizophrenia. However, a recent study found genetic overlap between schizophrenia risk genes and antipsychotic target genes, which suggests the pharmacological mechanisms may be polygenic and may also involve pathways that are not yet identified (
12).
Most genotype-phenotype relationships arise from complexity of cellular interactions (
13). Risk genes of polygenic disorders do not operate in isolation but in combination and interaction with other risk genes. The effect of a perturbation in one gene can propagate to affect other nearby proteins in the protein-protein interaction network, referred to here as the interactome. Therefore, protein products of genes that are associated with a particular disease tend to interact with one another and converge on related biological and functional networks (the so-called disease module) rather than being randomly spread throughout the interactome (
14,
15). Thus, network biology (
16) or network medicine (
15) provides an important framework where knowledge of protein-protein interactions can be used to gain more comprehensive insight into the molecular mechanisms of complex diseases (
14,
17).
The network approach also provides a unique opportunity to study drug effects by integrating the human interactome with knowledge of drug targets (
18). The Drug-Gene Interaction Database (DGIdb) (
11,
19) contains information on genes whose products are known to interact with drugs in humans (drug target genes), as well as genes that belong to the “druggable” genome (
11,
20). Okada et al. (
21) examined how targets of rheumatoid arthritis drugs map to the interactome neighborhood of identified risk genes for rheumatoid arthritis, and they found an overlap between drug target genes and risk genes as well as interactome neighbors of risk genes (
21). Another study examined how drug target genes overlap with GWAS hits across a variety of diseases, and it found very little direct overlap (
22). However, drug target genes showed threefold enrichment among the closest interactome neighbors, and enrichment was also significant among the second neighbors, suggesting that neighboring genes in the interactome should be included when searching for suitable candidates for drug repurposing (
22).
Here, adopting a novel network biology approach (
14), we study the schizophrenia disease module and its intersection with current antipsychotic drug target genes integrating information from the human interactome (
14) with data from the DGIdb. Our objective was to inform development of new medications (
20) by improving understanding of disease etiology and the function of current medications, as well as by identifying genes worth further examination as new drug targets or drug repurposing opportunities. We first identified and characterized a schizophrenia disease module in the interactome, and then we examined the interactome link between antipsychotic drug targets and schizophrenia risk genes through protein interactions. We revealed the specific risk genes and pathways that are involved in this link as well as the risk genes that are not linked to current antipsychotics. This information may be useful for identifying targets of future drugs that may also treat symptoms of schizophrenia other than psychosis.
Discussion
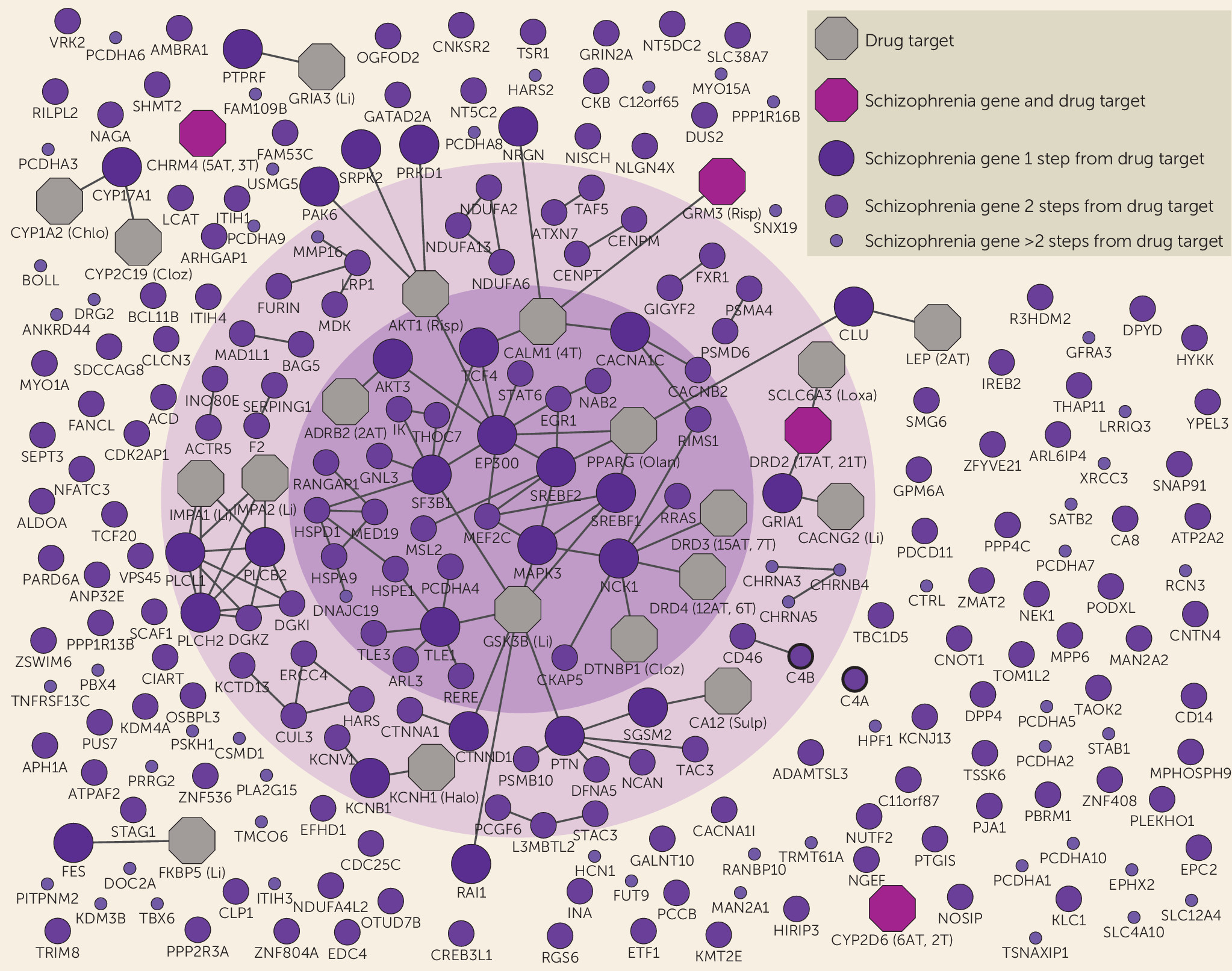
We examined the interactome link between antipsychotic drug targets and schizophrenia risk genes identified from the latest schizophrenia GWAS (
10). First, we identified a schizophrenia disease module, characterized by core genes involved in developmental biology and cognition. Second, we found antipsychotic drug target genes, as well as their first interactome neighbors, to be enriched for association with schizophrenia. Through network graphs, we found the observed interactome link between existing drugs and the disease to be located among core genes in the disease module and to involve multiple pathways. Important risk genes that were not linked to current drug targets were also identified.
Using network methods and a high-confident human interactome (
14), we showed that schizophrenia risk genes are significantly localized rather than randomly scattered in the interactome, forming a distinct disease module (
14). Previous pathway analyses performed on schizophrenia risk genes from the latest large-scale GWAS (
10) treated all risk genes equally and mainly identified enrichment for synapse- and dendrite-related pathways (
33). One problem with the previous approach is that many GWAS-identified genetic loci contain several genes, of which all may not be causal to the disease. Importantly, it has been shown that when several genes are present in the same loci of a GWAS, risk genes that interact with other risk genes are more likely to be causally linked to the disease (
34,
35). Restricting analyses to interconnected risk genes enabled the identification of novel and highly disease-relevant biological processes and pathways (see Supplementary Table 1B and 1C in the
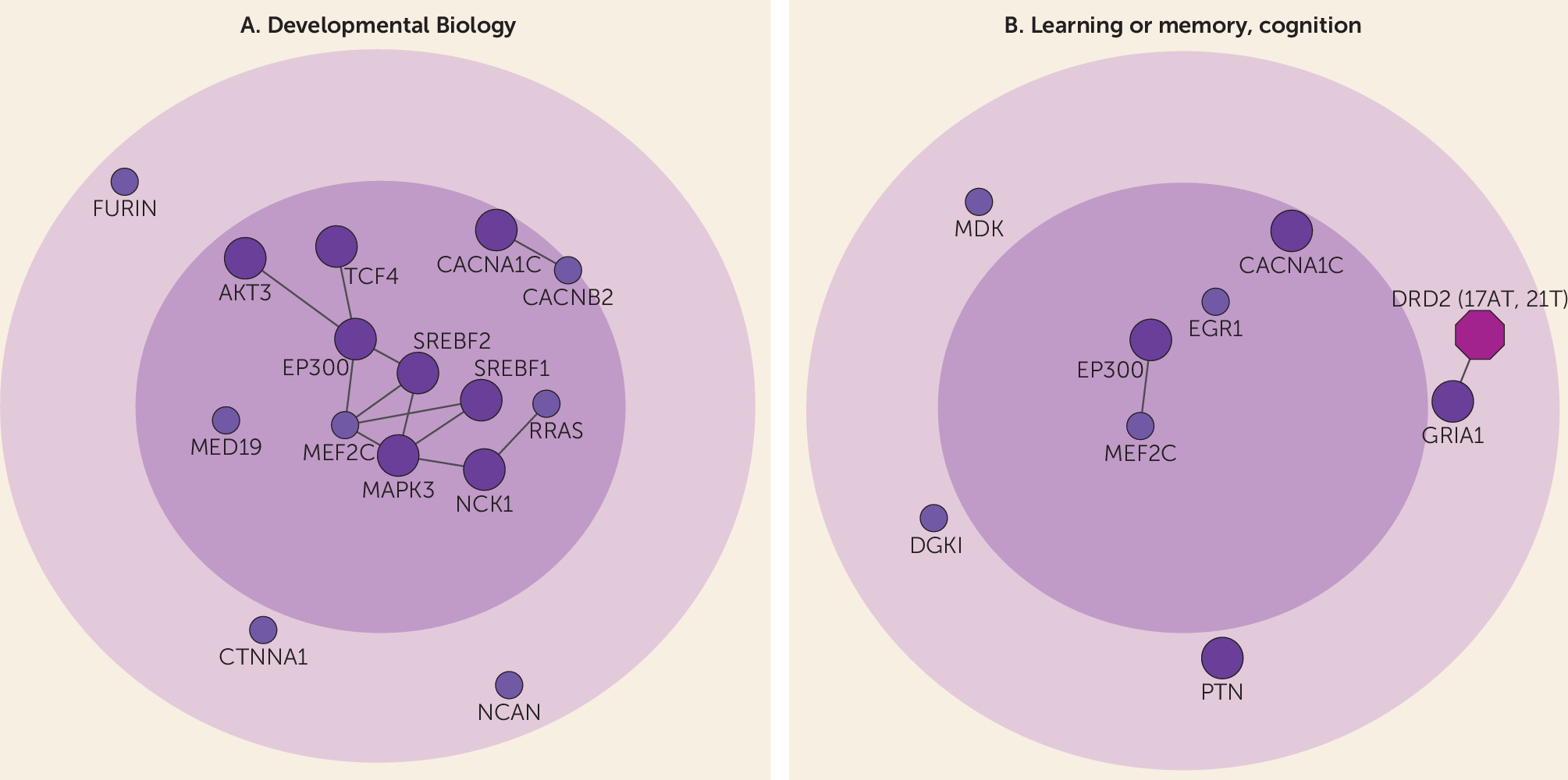
online data supplement). These core genes were most strongly related to developmental biology, which may constitute a central role in disease etiology (
Figure 2A). Interestingly, core genes were also enriched for cognition-related processes and pathways (
Figure 2B). Although schizophrenia is diagnosed based on positive and negative symptoms, cognitive impairment is a core clinical feature of the disease, with evidence of premorbid cognitive impairment from as young as age 7 (
36).
We confirmed previously reported enrichment of antipsychotic drug target genes for association with schizophrenia (
12) and, importantly, also found enrichment for neighbors of antipsychotic drug target genes. These results indicate that the pharmacological mechanisms of current antipsychotics overlap with the pathogenesis of schizophrenia and that the drug-disease link extends beyond dopamine to also involve other neurotransmitter systems, such as glutamatergic and cholinergic pathways (
Table 1; see also Supplementary Table 2B in the
data supplement). These pathways not only include risk genes directly targeted by antipsychotics (the glutamatergic and cholinergic receptor genes
GRM3 and
CHRM4) but also risk genes interacting with drug targets, such as
CACNA1C, coding for a calcium channel subunit, and cell signaling kinases such as MAP kinase 3 (
MAPK3) and AKT serine/threonine kinase 3 (
AKT3). Indeed, previous efforts have identified potential drug targets within the cholinergic and glutamatergic systems (
37), and pharmacogenetic studies have linked antipsychotic response to variants in
GRM3 (
38). Genetic variations in
CYP2D6, a gene linked to drug metabolism and one of the schizophrenia risk genes directly targeted by antipsychotics, have been linked to side effects of antipsychotics (
39). Some studies also linked variants in
CYP2D6 to antipsychotic response, but results have been mixed (
40). Although we did not observe significant overlap between antipsychotic targets and control conditions in our enrichment analyses, it remains possible for partial overlap at the individual gene level (e.g., pleiotropic variants), and this warrants future investigation.
Although genes close to current antipsychotics were related to learning and cognition, antipsychotics show only weak improvements of cognition in randomized controlled studies (
41). Most antipsychotics reduce dopamine release, although the differential direction of altered dopamine levels in different brain areas has been linked to different aspects of schizophrenia (
42–
44). Dopamine dysfunction cannot explain all aspects of schizophrenia, and to develop improved medication, it is important also to focus on other potential targets. We identified the risk genes that are least connected to current antipsychotics together with their first neighbors (see Supplementary Figure 1 in the
data supplement). This information may be used together with other sources of knowledge, such as the druggable genome (
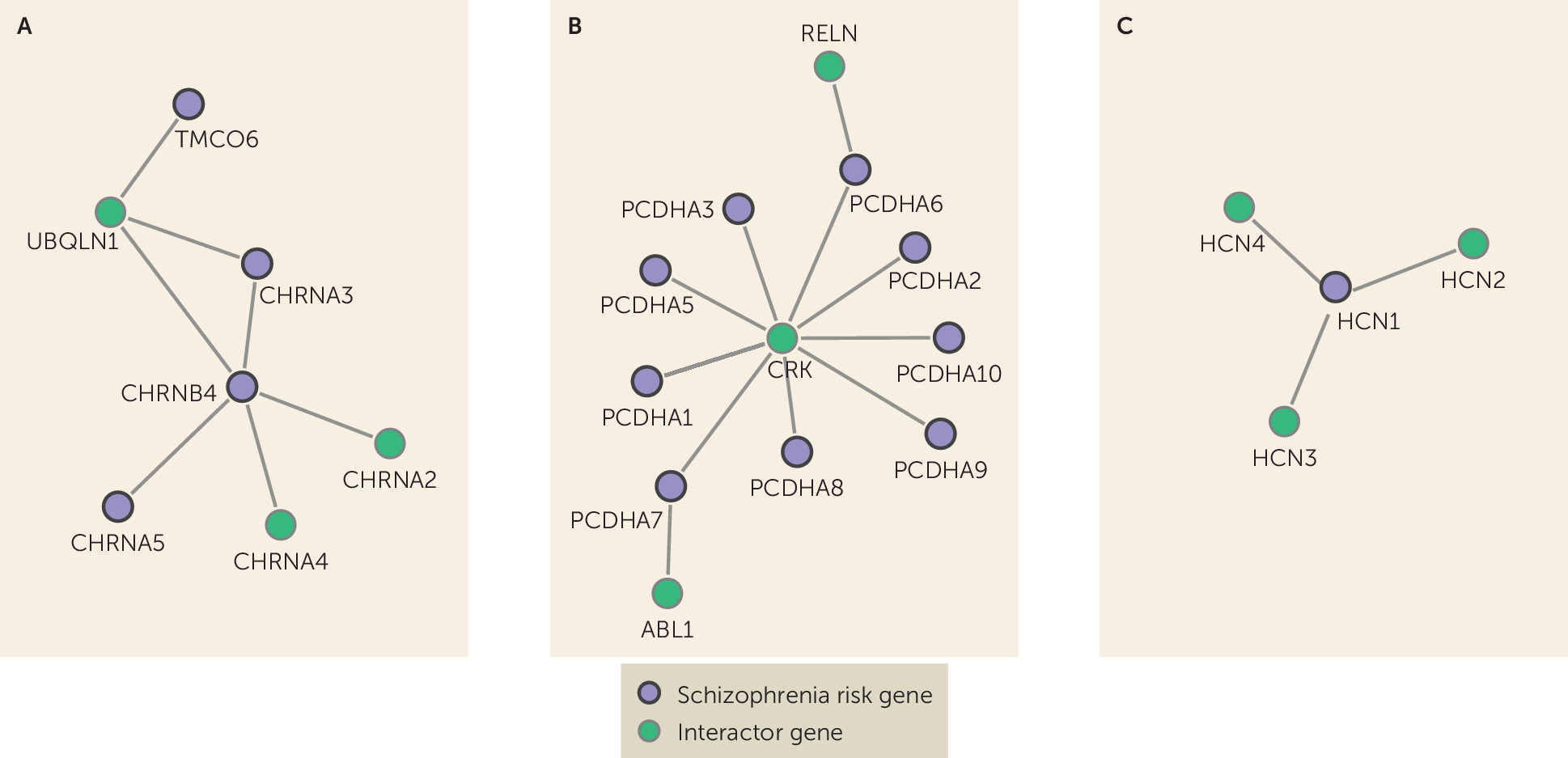
11), to inform future studies aiming to identify candidate targets for new drugs or drug repurposing to treat symptoms that are little affected by current antipsychotic medication, such as cognitive-enhancing drugs. Among these genes, important examples are nicotinic acetylcholine receptor genes (nAChRs) (
Figure 3A). The nAChRs are thought to be important for the cognitive symptoms of schizophrenia (
45) and are also implicated in Alzheimer’s disease (
46,
47). The nAChRs are targeted by several existing drugs, including the Alzheimer’s disease drug galantamine (see the
data supplement). Galantamine has been studied in humans with schizophrenia as well as in animal models, with indications of improved effect on cognitive symptoms and enhanced efficacy of antipsychotics in rats (
48,
49). The nAChRs are also currently being examined as potential drug targets to enhance cognition in schizophrenia patients (
50), further justifying our hypothesis that suitable drug targets may be found among these genes. Other interesting genes that did not overlap with antipsychotic drug genes are the Protocadherin gene cluster (
PCDHA1–10) and hyperpolarization activated cyclic nucleotide gated potassium channel 1 (
HCN1) (
Figure 3B and
3C). The
PCDHA genes are neural cell adhesion proteins involved in neural differentiation during development (
51), processes thought to be important for the development of schizophrenia. Protocadherin genes have also been linked to cognition, personality, and mood disorders (
52). The
PCDHA genes are not considered druggable (
11), but two neighbor genes are (
RELN and
ABL1).
HCN1 is druggable (
11) and may have a link to cognition in schizophrenia through involvement in mechanisms of synaptic plasticity and memory (
53). Furthermore, these ion channels have been suggested as potential new targets for depression (
54) and cognitive dysfunction in neurofibromatosis type 1 (
55).
Limitations
A major limitation of the human interactome is potential bias toward well-studied proteins. To minimize this bias, the Menche et al. interactome also includes protein-protein interactions derived from unbiased high-throughput data sets (
56–
58), and we found that schizophrenia risk genes were significantly localized also when using node degree–preserved methods. Also, the known human protein-protein interaction network covers only an estimated 20% of all potential protein-protein interactions (
14). However, the current level of network completeness was shown to successfully identify the disease module of 226 complex diseases (
14). The available drug target data are also incomplete. Among the identified risk genes with more than two steps to a drug gene target, some may have undiscovered connections with drug target genes. To further address this limitation, we repeated our analyses using the less conservative STRING interactome, with consistent results.