By leveraging RNA sequencing and genomic data from a large sample of DLPFC tissue from completed suicide cases, nonsuicide patients, and healthy control subjects and a focus on the suicide method, we identified signatures associated with suicide death and aspects of its phenomenology. We show that those who die by suicide by violent methods may have a separate clinical condition from other suicide cases and from nonsuicide psychiatric patients with similar diagnoses. G protein-coupled purinergic signaling may play a role in this distinction and in related aggressive phenotypes, in a specific mitochondrial background of altered energy metabolism.
A previous attempt at genome-wide expression profiling in suicide via next-generation sequencing (
58), not taking into account means of suicide, failed to detect FDR-significant effects specific to suicide and not to major depression, likely because of limited sample size (total N=59, of which 21 were cases of suicide). That study, however, suggested potential deficits in microglia and ATPase activity in suicide. Additionally, an earlier microarray investigation (
59), comparing suicides with nonsuicides with the same diagnosis (schizophrenia), found up-regulation of purinergic signaling (including
P2RY13) in suicide, as did a recent study (
60) looking at a selected pool of glia-related genes (including
P2RY12) using real-time qPCR. By studying a larger sample with RNA sequencing, we have been able to parse out the effect of suicide on the brain transcriptome in the context of a psychiatric diagnosis and highlight the signaling involved, while providing novel insight into this extreme behavior.
Suicide by Violent Means May Be a Distinct Condition
The possibility that suicide by violent methods is a biologically meaningful designation—that is, relatively orthogonal to conventional psychiatric disorders—is first suggested by our analyses of gene expression. We showed that in the patients who died by suicide by violent means, the expression patterns are different not only from other patients diagnosed with the same psychiatric disorders but also from patients who died by suicide by nonviolent means.
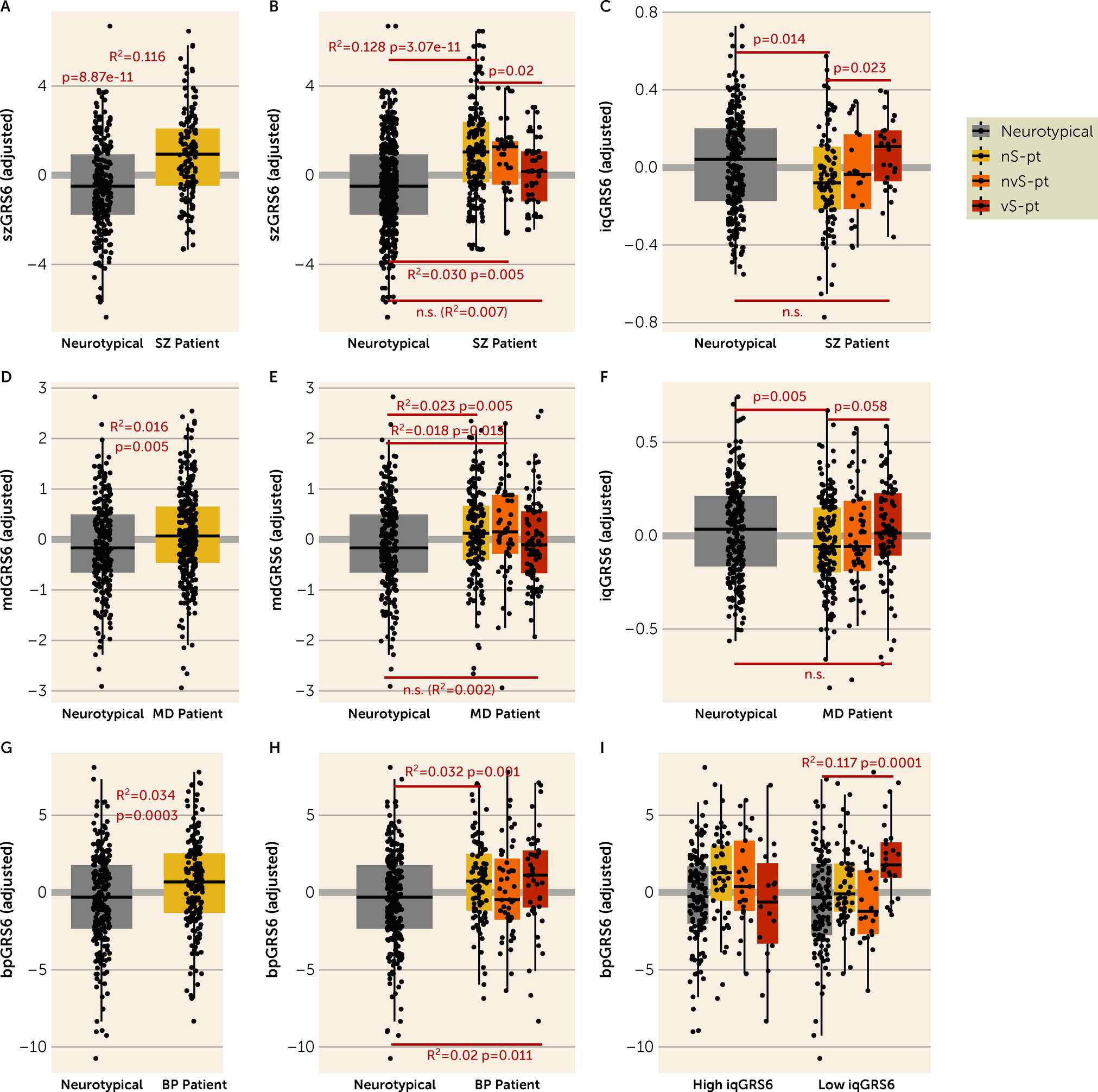
We also investigated genomic risk, which is less likely to be confounded by the experience of the psychiatric condition or by postmortem tissue artifacts. The GRS analyses largely mirror the transcriptomic findings that patients who died by suicide by violent means show only marginal, if any, differences from neurotypical individuals in genomic risk, as well as divergence from nonsuicide patients and nonviolent suicide patients in genomic risk for their clinical diagnoses. Patients affected with schizophrenia or depression who died by suicide by violent means are less “genetically susceptible” to their diagnosed disorder, and more “genetically inclined” to higher IQ than other patients. Additionally, patients affected with schizophrenia who died by suicide by violent means are also more “genetically resilient” to their diagnosed disorder. These results support the conclusion that individuals with a diagnosis of schizophrenia or depression who select a violent means of suicide are distinguishable biologically from similarly diagnosed patients who do not make this extreme choice.
In contrast to the findings in patients diagnosed with either schizophrenia or depression, violent suicide patients with a diagnosis of bipolar disorder do not differ in diagnosis GRS compared with other patients with the same diagnosis. That those patients evince genetic risk more in line with others with bipolar disorder may be explained by evidence that this diagnosis is highly associated with impulsive behavior and with suicide, with suicide rates in some studies up to 20 to 30 times higher than in the general population (
61). However, the possibility that, in general, violent suicide is associated with higher IQ GRS and low diagnosis GRSs is also supported by the analysis in patients with bipolar disorder, where, in the presence of high IQ GRS, violent suicide patients have lower bipolar disorder GRS compared with other patients.
Perhaps surprisingly, the genomic profile of the patients who died by violent suicide is not associated with suicide attempt GRS, which is consonant with the notion that genetic studies of suicide attempt may not catch the genetic architecture of the actual completed behavior, especially by violent methods. This is likely because they include, by design (
43), people who survived a suicide attempt, thus excluding the more than 50% of suicidal patients who die in their first attempt (
56) and who may better represent those at highest genetic risk for suicide completion (
62).
The Potential Role of Purinergic Signaling and Mitochondrial Function
Our molecular data further suggest that suicide by violent means, while less biased by genetic risk for conventional psychiatric disorders, may share characteristics with entities such as aggression, as supported by the aggressive phenotype in
Drosophila and by our previous findings in humans (
14). Indeed, the DE analysis implicates purinergic signaling, and genes linked to aggressive behavior in two GWAS analyses in
D. melanogaster (
20) also point to purinergic-related terms as the most enriched processes. The purinergic DEGs belong to the coexpression network studied in our previous publication (
14), with
P2RY13 being the top gene most strongly related to
LINC01268.
P2RY13 codes for a transmembrane receptor, enriched in microglia (
63) and involved in microglia-mediated hippocampal neurogenesis (
64). Included in the same network is also
P2RY12, a purinergic DEG and microglia-enriched gene as well (
63), whose knockout produces behavioral alterations in mouse brain (
65). P2Y12 receptors are involved in lithium pharmacodynamics (
66), an interesting notion in light of lithium’s antisuicide properties. P2Y12 receptor signaling is also required for physiological microglia-neuron communication at somatic junctions (
67,
68); notably, other human microglia signatures appear at the top of violent suicide DEGs, including
CX3CR1,
TMEM119, and
SELPLG.
Intriguingly, the potential involvement of purine signaling in suicide by violent methods and in aggressive behaviors resonates with aspects of the syndrome caused by mutations in the gene
HPRT1—Lesch-Nyhan disease (
69). The most defining behavioral manifestation of the disease is the irresistible impulse to self-injury, including lip and finger biting, eye poking, and head banging (
69). The etiopathogenesis of Lesch-Nyhan disease is poorly understood; however, as a result of
HPRT1 deficiency, purine recycling to their nucleosides is not functional, leading to enhanced de novo purine synthesis (
70). Of note, a recent study has shown that in mouse brain and plasma, prophylaxis with ketamine following stress is associated with long-term alterations of purine and pyrimidine metabolism (
71); ketamine is also known to transiently reduce suicidal ideation with a single dose (
72).
We searched for upstream regulators of the DEGs in violent suicide patients compared with nonsuicide patients, and compared with nonviolent suicide patients, and the analysis pointed to
EIF4E, a critical regulator of translation. Intriguingly, a recent study has found that cell-specific translation via eIF4E is central to the antidepressant activity of ketamine (
73). According to these data, ketamine would exert its therapeutic action by activating eIF4E; consistently, the directionality of our DEGs would suggest an upstream inhibition of this translational regulator.
Among violent suicide DEGs, the most significant is the functionally unknown
LINC00996, which flanks several GTPases, IMAP family genes (GIMAP), some of which are also violent suicide DEGs. GIMAP genes in this region have previously been linked to completed suicide cases within high-risk families, regardless of co-occurring psychopathology (
48).
LINC00996’s expression is also restricted to microglia (
74); importantly, an independent bioinformatic investigation (
75) showed G protein-coupled purinergic nucleotide receptor signaling as one of the top pathways enriched for the genes related to
LINC00996.
The DE results posed the challenging question of how the relatively fewer biological differences between violent suicide patients and neurotypical individuals identified here translate into their markedly different behavior. We cannot rule out the possibility that more substantial differences will emerge in transcriptional analyses of other brain regions, or in single cellular phenotypes, which are the subject of future investigation, or at epigenetic and proteomic levels, consistent with the enrichment of upstream signal to DEGs in violent suicide for translation regulators such as
EIF4E. Meanwhile, the DE based on this contrast highlights only eight DEGs, the top of which is
ELFN2, a recently identified adhesion molecule that selectively binds metabotropic glutamate receptors.
Elfn2 knockout mice show susceptibility to seizure, anxiety/compulsivity, and hyperactivity (
76). However, this gene is not DE in violent suicide patients compared with nonsuicide patients, which suggests that the result may be driven by case-control status.
According to the linear model results, neurotypical individuals are in the middle of a continuum of gene expression, featuring the purinergic DEGs, with the two extremes being violent suicide and nonsuicide/nonviolent suicide patients. One possible explanation for this evidence is that purinergic signaling functions optimally within a narrow window of expression, where lower expression is associated with disease status, and higher expression to health, but only within a certain threshold, as has been reported for other genes linked to neurotrophism (
77).
Interestingly, the WGCNA results suggest that in the context of suicide by violent means, the purinergic DEGs may function in an opposite state of energy metabolism to that observed in all other individuals. Indeed, purinergic DEGs tend to be more coexpressed in the context of higher energy metabolism in the violent suicide cases, in contrast to lower energy metabolism coexpression in neurotypical individuals and the other groups. As mentioned, recruitment of microglial processes to somatic junctions is linked to mitochondria neuronal activity in neuron-microglia communication (
67,
68). Recent evidence (
78,
79) has further outlined a key role for microglia in neuronal inhibition, by converting ATP to ADP, which stimulates P2Y12 microglia receptors. In this light, another finding of the WGCNA data is the association with violent suicide of a module enriched for GABA synthesis. Thus, our data point to violent suicide, in contrast to the other three conditions, as involving a state of excessive neuronal-microglia communication that may be detrimental to brain function.