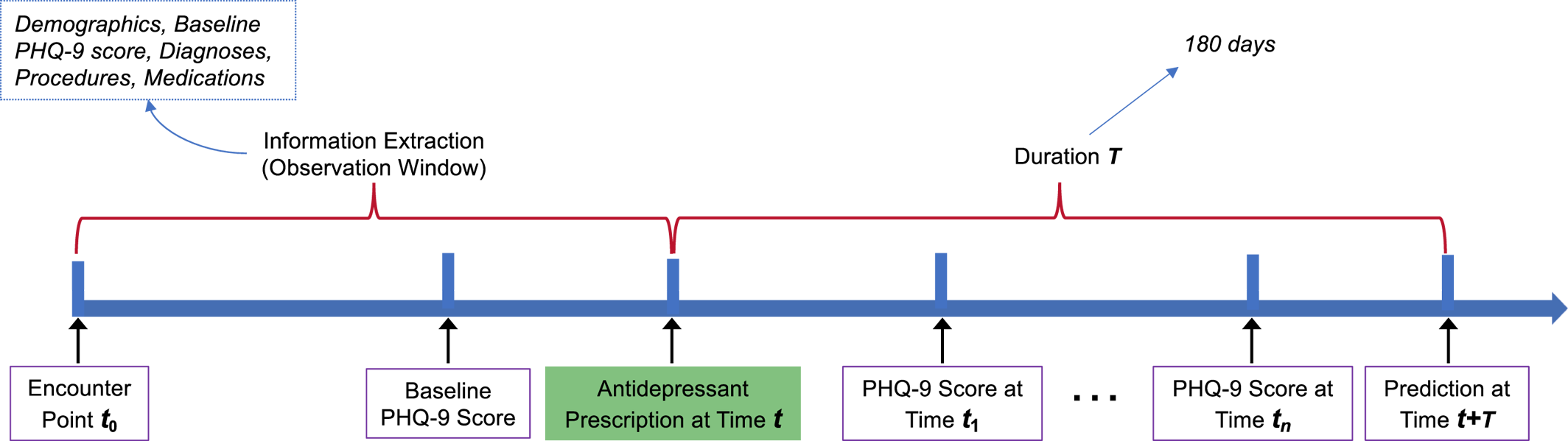
Depression is one of the most prevalent psychiatric disorders, affecting approximately 14% of the global population (
1). The economic costs resulting from depression are staggering and have become the second contributor to disease burden (
2). While antidepressants are commonly prescribed to patients suffering from depression (
3), due to the complex etiology and heterogeneous symptomatology of depression, prior studies suggest that antidepressant treatment efficacy is usually low, with as few as 11–30% of depressed patients obtaining remission after initial treatment (
4). The use of prediction tools in areas of medicine such as oncology, cardiology, and radiology has played an important role in the clinical decision‐making (
5,
6), suggesting the potential utility for such tools in predicting antidepressant treatment efficacy.
Recent studies have assessed the prediction of antidepressant treatment outcomes such as responder or the achievement of remission using brain images, social status, and electronic health records (EHRs). In particular, in studies using functional magnetic resonance imaging (fMRI) data (
6,
7,
8,
9), the mean activation and differential response were analyzed for case and control groups, and specific observations were found to be predictive of the response to antidepressant treatment. However, the cost and time associated with collecting and processing fMRI data may hinder the use of the approach in a practical manner (
4). Previous studies also used patient self‐report data, including socioeconomic status (
4,
10,
11), to evaluate whether patients would achieve symptomatic remission. However, self‐reported social information may be less precise as an outcome measure and prone to nonresponse bias.
In addition, prior studies based on EHRs mainly extracted feature information from patients' medical records (
10,
12,
13,
14), such as medication dose information, to predict treatment dropout or remission after receiving antidepressants. However, these studies did not consider patients' baseline depression severity and other clinical data such as diagnostic codes in prediction model development. Furthermore, most of the previous studies defined the outcome based on a behavioral assessment summarized in a numerical variable (e.g. Montgomery‐Asberg Depression Rating Scale (MADRS) score or Patient Health Questionnaire‐9 (PHQ‐9) score) (
6), which only considered the baseline and final scores of a specified treatment period, without taking the variations in patients' self‐report scores and severity of depression over that time interval into account.
DISCUSSION
This study demonstrates the potential of using machine learning to identify clinically meaningful predictors of the outcome of antidepressant treatment, especially when a slope fit of all PHQ‐9 scores represents longitudinal treatment recovery. In the investigation of 808 individuals with EHRs, predictive models including LR, RF, and GBDT were built for discriminating outcome by integrating multiple types of clinical information such as demographics, diagnostic codes, and medications. Combining multiple types of features could build more complete representations of patients, which can improve the predictive performance of ML models.
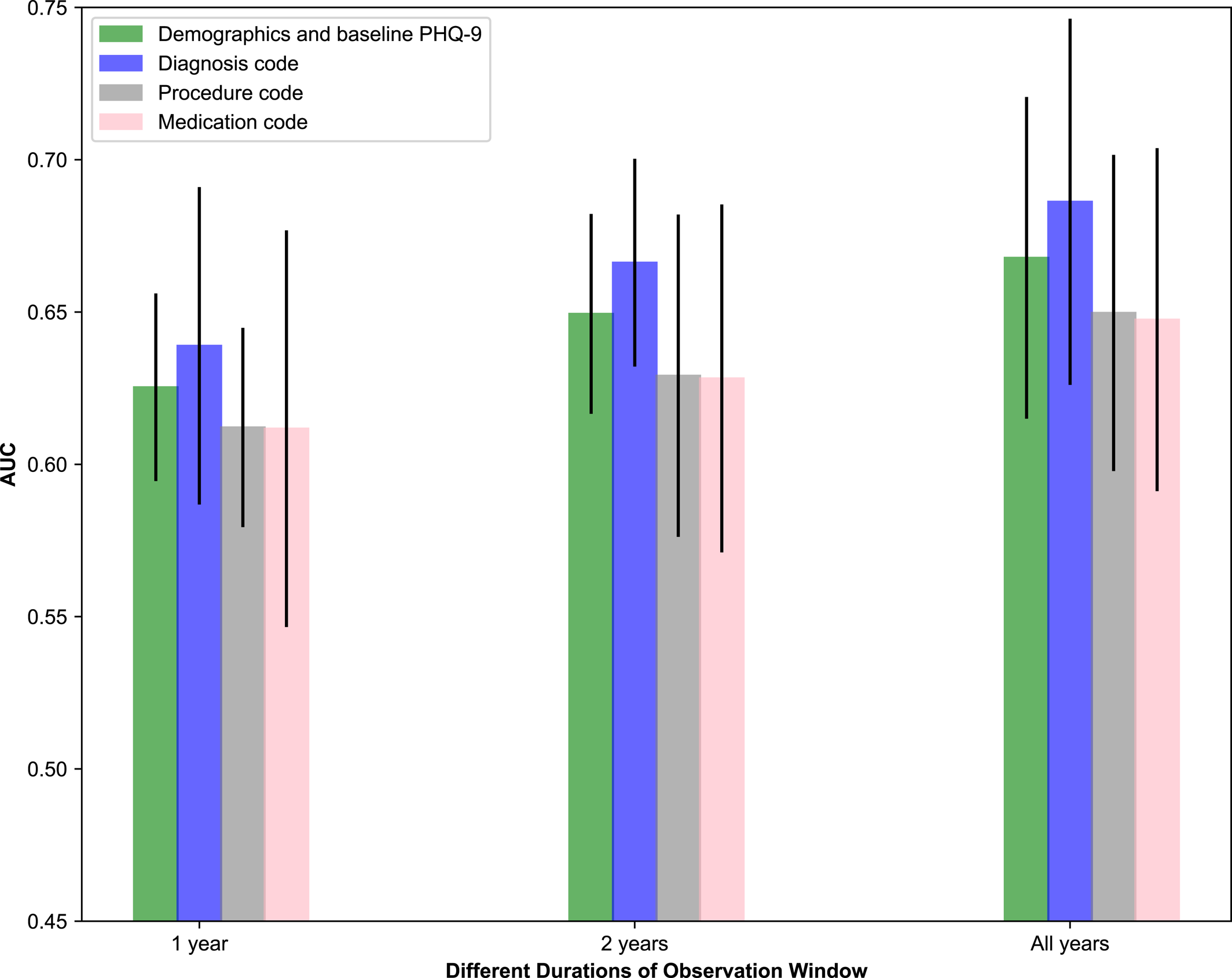
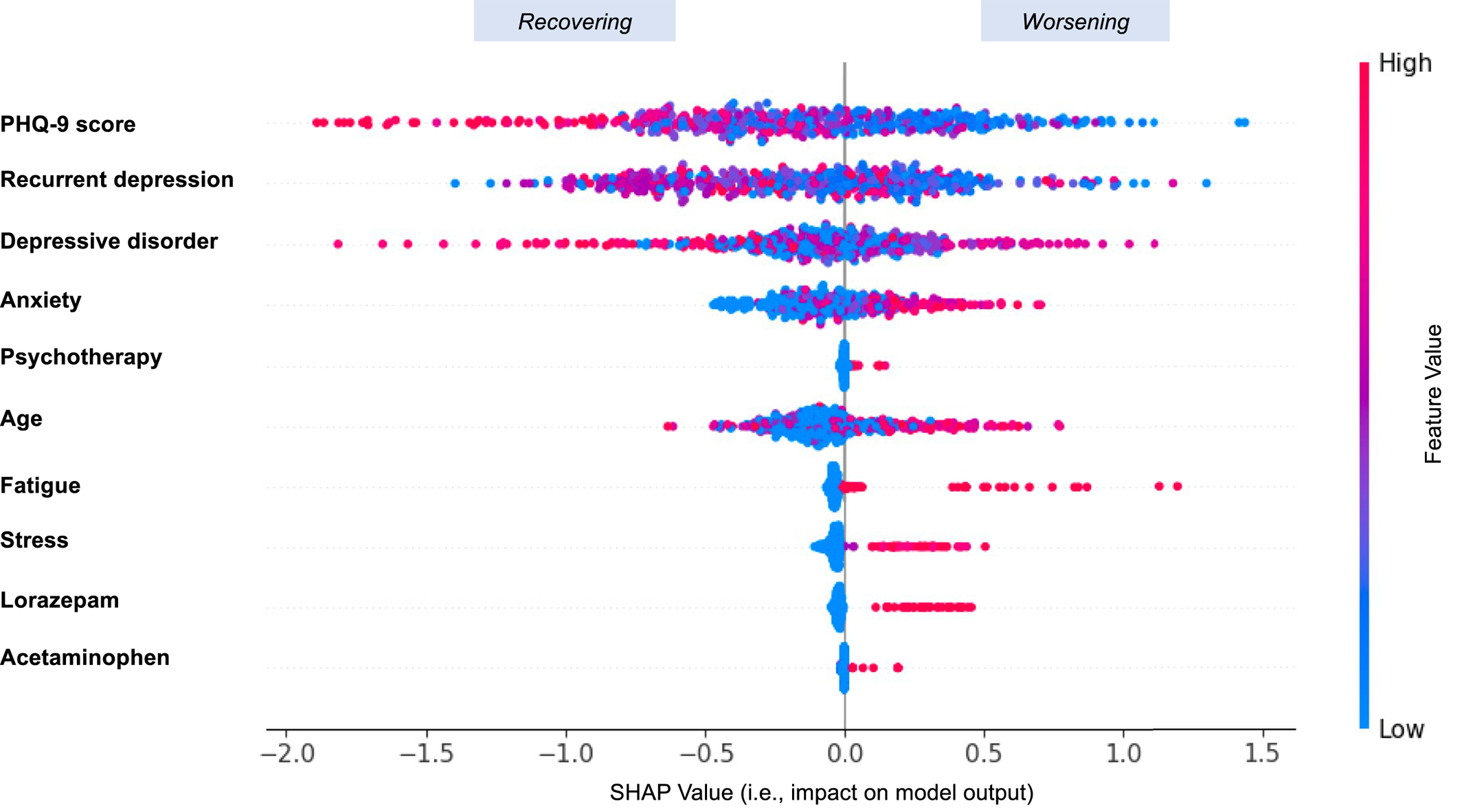
In this study, the discrimination was modest, with an AUC of 0.7654 (SD: 0.0227) obtained by GBDT when considering EHRs from “all years.” Discrimination in these models differed when considering 1 year, 2 years, and all years worth of clinical data, and we observed that considering more longitudinal data tended to improve prediction performance. By removing individuals with low PHQ‐9 scores (<10) at baseline, a modified AUC of 0.7254 (SD: 0.0218) was obtained by GBDT. This resulted in a large drop in AUC, because the AUC measures how well the predictive model can distinguish patients who recovered from those who worsened. It is reasonable to expect the exclusion of patients with low baseline PHQ‐9 scores to limit the model's ability to distinguish worsening individuals because it is unusual for patients with severe baseline scores to worsen further after treatment initiation. In this sense, a broader range of PHQ‐9 scores may hold more potential for improved distinction. We also observed that different types of clinical information played different roles in prediction. Diagnostic information such as anxiety, psychotherapy, and recurrent depression contributed the most to prediction. Demographic and baseline PHQ‐9 score information combined played a more important role relative to procedure and medication codes separately, which may be because baseline PHQ‐9 score was a vital contributor on its own. These findings could corroborate well with a previous report (
15). In addition, if a patient was more complex and had more severe comorbidities such as anxiety and stress prior to taking the antidepressant, the model tended to predict the outcome as “Worsening.”
Our study differs substantially compared to previous work that investigated antidepressant treatment outcome (
6,
10,
12,
28,
29,
30). To the best of our knowledge, no prior work has attempted to model antidepressant treatment outcome based on the slope of multiple continuous self‐report PHQ‐9 measurements over time, and very limited studies have utilized complete, longitudinal EHRs for predicting antidepressant treatment outcome. Previous literature only considers the difference between the final PHQ‐9 score and the baseline PHQ‐9 score for a certain treatment period (
6,
10,
30,
31). Relying on these two timepoints alone can be misleading because the single difference in scores might suggest a worsening or improving outcome, when in fact the course of the outcome was mostly in the other direction. Our work fills an important gap by using the slope fit using a linear regression model based on all PHQ‐9 scores throughout a certain treatment period. This method is advantageous because it captures the evolution and intermediate oscillations in PHQ‐9 scores over time towards modeling the overall treatment outcome.
Pradier et al. attempted to predict treatment dropout after antidepressant initiation using EHRs (
12). In their study, the primary outcome was treatment discontinuation following index prescription, defined as less than 90 days of prescription availability and no evidence of non‐pharmacologic psychiatric treatment. However, treatment discontinuation may not necessarily reflect a “recovering” or “worsening” treatment outcome and moreover is unable to account for variation among patients in terms of depression severity at the time of treatment. Common measures such as PHQ‐9 and HDRS (Hamilton Depression Rating Scale) are more robust in investigating antidepressant treatment outcome (
15,
30), because they are acquired based on standardized questionnaires and provide more complete and objective information for estimating antidepressant treatment outcome (
32).
Our study has important clinical implications. First, the machine‐learned class models predicted antidepressant treatment outcome using patients' medical history, which may encourage clinicians and patients to conduct more follow‐up visits during the course of their treatment (
12). Additionally, the predictive results obtained from the models can aid clinicians in developing treatment plans that combine multiple elements in sequence or in parallel. Furthermore, the predictive models using EHRs can make contributions to personalized treatment management strategies in psychiatry (
33). Beyond informing targeted treatments, these predictive models may potentially contribute to the design of a new generation of EHR‐linked clinical trials (
34). For example, clinicians can stratify the patients into “high‐risk” and “low‐risk” groups based on predictive results (“worsening” or “recovering”) and pay closer attention to the treatments and prognosis of the “high‐risk” group (
35).
There are several potential limitations to our study. For example, this study only considered data from a single academic medical center, which did not allow us to generalize the model prediction across multiple different health systems. This may have also resulted in a sample that was selective in geography, payer, and patients, and may not fully represent the population at risk. As with most EHR‐based studies, we are limited only to visits captured within the EHR network, and as such, clinical care sought outside this network may be missing. Also, when defining the outcome using the slope, the slope of zero may not always be the clinically meaningful cut‐point for recovering or worsening depression, as reversions to mean would be expected. Further investigation is necessary as different thresholds may produce different “Recovering” and “Worsening” distributions, which would present different prediction performances. In addition, it was unknown which diagnosis a given patient was referred or receiving treatment for, and we did not build predictive models for different classes and doses of antidepressants in this study. Accounting for different antidepressant classes, extending the study period to explore longer term outcomes, as well as conducting cross‐site validation may further enhance the results and applicability of this study, which will be investigated in future studies. Subsequent studies may also apply the techniques used in this analysis to information used in previous studies that investigated antidepressant outcome prediction, for example, socioeconomic status for which objective measures and structured EHR data are expanding. Additionally, natural language processing techniques would be considered in the future to process clinical text. With these limitations in mind, these results provide insights in terms of personalizing antidepressant treatments and encouraging researchers to pursue modeling of this simple but highly valuable outcome.