Psychiatric pharmacogenomics is defined as the assessment of genomic variability to clinically inform the selection and dosing of psychotropic medications. Genetic testing of pharmacogenomically informative genes has only been available to clinicians in recent years. The rapidly expanding number of genes that are now being tested in clinical laboratories is in large part the consequence of a considerable decrease in the cost of genetic testing. As the availability of pharmacogenomic testing increases, it is reasonable to expect that patients will have fewer side effects and will therefore adhere more readily to treatment recommendations. The goal of this review is to provide a synthesis of recent advances in the field of psychiatric pharmacogenomics. This will include a discussion of the genes that are currently being tested. However, these initial genotyping opportunities represent only the very first examples of how results of genetic testing will be able to inform psychiatric treatment. Pharmacogenomic genotyping to guide the selection and dosing of antidepressant medications is described in this review. However, the principles and applications of pharmacogenomic genotyping are equally relevant for other classes of psychotropic medications, including antipsychotic medications, stimulants, and mood stabilizers.
There are two primary reasons for psychiatrists to order pharmacogenomic testing. The first objective is to minimize the occurrence of adverse reactions to psychotropic medications. Given the appropriate national commitment to improving the safety of psychotropic medication management, it is not surprising that the initial utilization of genetic testing has been to minimize side effects and improve the overall safety profile of the use of these medications in clinical practice. However, the second objective, which is ultimately the more far-reaching of the two goals of pharmacogenomics, is to identify medications that will have the highest probability of efficacy for an individual patient. Given that many available psychotropic medications exert their therapeutic benefit through a range of mechanisms and that these antidepressant medications are metabolized by different hepatic enzymes, the potential impact of pharmacogenomic testing for the treatment of depression promises to be substantial.
The clinical significance of variations in genes coding for the enzymes that play a major role in drug metabolism has been recognized for many years. For the most part, variations in the structure and functions of the genes that code for these enzymes have had an impact on the pharmacokinetics of their substrate medications. These genes are often referred to as “drug-metabolizing enzyme” genes or more simply as DME genes (
1).
Genes can also influence the impact of medications on patient response by changing how receptors function or how neurotransmitters are transported. These genes that code for receptors or transporters are generally referred to as “target genes.” Structural or functional variations in these genes result in a pharmacodynamic influence on medication response.
Tests for several of the DME genes are now routine, and the results can be used to predict pharmacokinetic phenotypes. Two of these DME genes, 2D6 (i.e., the cytochrome p450 2D6 gene) and 2C19 (i.e., the cytochrome p450 2C19 gene), will be reviewed. More recently, the genotyping of target genes has become available for use in the clinical management of patients. Two genes coding for proteins that play important roles in serotonin neurotransmission, SLC6A4 (i.e., the serotonin transporter gene) and HTR2A (i.e., the serotonin 2A receptor gene), will also be reviewed.
The 2D6 gene
The 2D6 gene is located on the 22nd chromosome and has a high degree of allelic variability. This gene codes for the cytochrome P450 2D6 enzyme, which is involved in the primary metabolism of more than 70 medications. More than 100 variations or alleles of this gene have been recorded. Furthermore, the 2D6 gene has the relatively unique characteristic of having multiple copies of the gene occurring on the same 22nd chromosome. Multiple copies of the 2D6 gene can occur on the same chromosome as a consequence of the small size of the chromosome and the fact that it contains multiple homologous sequences. As a result, the 22nd chromosome has a high rate of “uneven crossover events” that take place during the process of meiosis. Individuals who have three or more active copies of the 2D6 gene are referred to as ultra-rapid metabolizers.
The identification and classification of many completely inactive and partially inactive alleles of the 2D6 gene has gradually taken place over the past 10 years. The primary goal of genotyping patients who are to be given an antidepressant has been to identify individuals who have two completely inactive copies of the gene. These individuals are described as poor 2D6 metabolizers, and they have a very low tolerance for virtually all 2D6 substrate medications (
2). However, there is another subset of slower metabolizers who have one inactive copy and one partially inactive copy. In this case, some tolerance of 2D6 substrate medications in low doses may be possible, although the risk of side effects in these individuals is increased. In recent articles, these individuals have also been referred to as poor metabolizers.
In some of the older scientific literature individuals who had one inactive copy of the 2D6 gene and one partially active copy had been referred to as “intermediate metabolizers.” More recently, the label intermediate metabolizer has been used to refer to individuals who have one active copy and either a totally inactive second copy or a partially inactive second copy (
3). Whereas intermediate metabolizer patients who have only one active copy of the 2D6 gene do have an increased sensitivity to 2D6 substrate medication in high doses, the degree of tolerance of a medication depends on the specific inactive allele that an individual carries. As a general rule, intermediate metabolizers can tolerate 2D6 substrate medications at low to moderate doses.
Individuals with two active copies of the 2D6 gene produce sufficient enzyme to tolerate virtually all 2D6 substrate medications at standard doses. These individuals are referred to as “extensive metabolizers.”
Commonly prescribed antidepressant medications can be categorized on the basis of the degree to which they are 2D6 substrate medications (Table 1). Medications such as paroxetine and venlafaxine are primarily metabolized by the 2D6 enzyme. In contrast, drugs such as sertraline and duloxetine have at least one other functional alternative pathway and would consequently be more suitable choices for individuals with compromised 2D6 metabolism. Other antidepressant medications such as escitalopram and fluvoxamine are minimally metabolized by the 2D6 enzyme and would be expected to be well tolerated by individuals regardless of their 2D6 genotype.
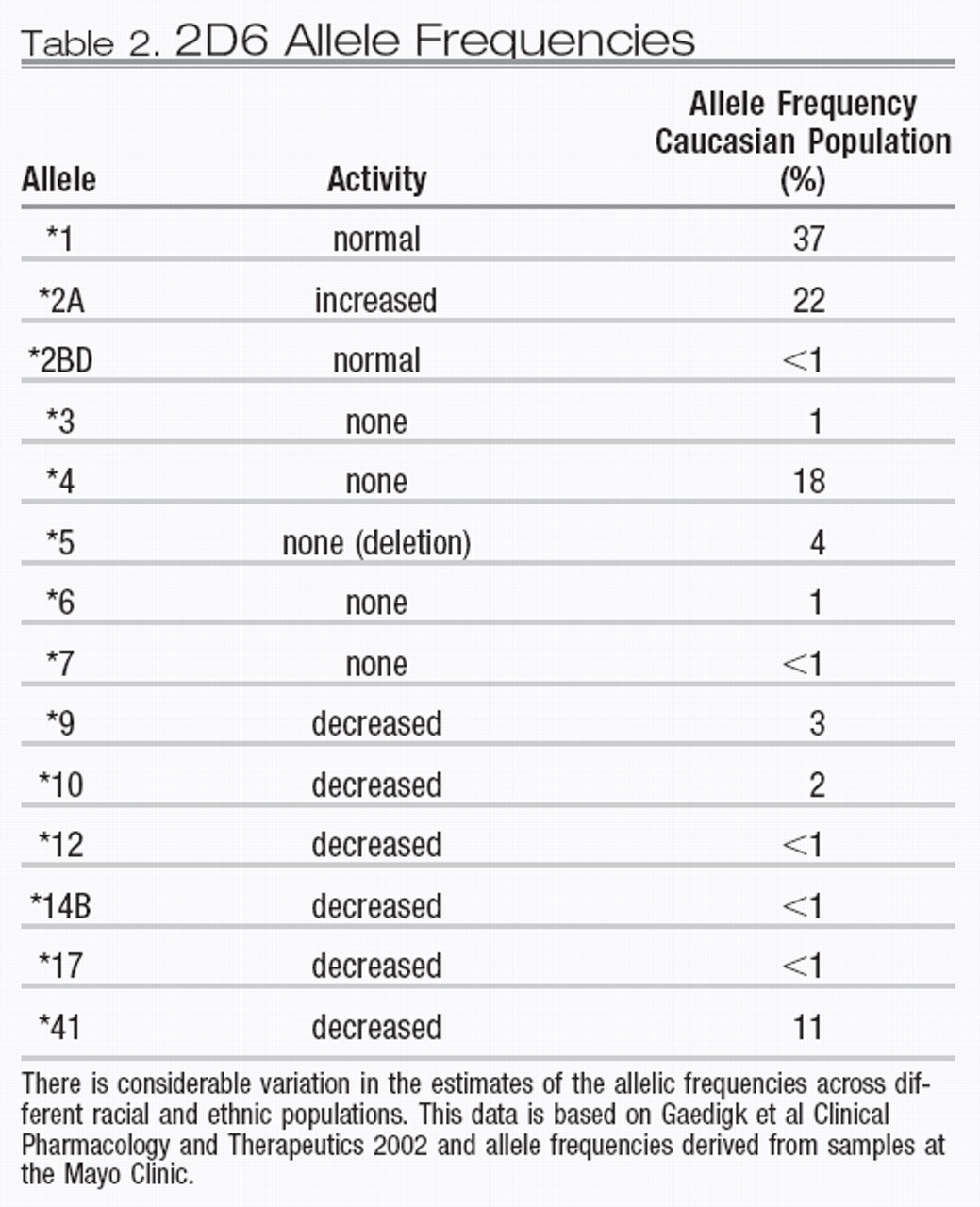
The frequency of 2D6 alleles varies in different ethnic groups. A distribution of the most common alleles in Caucasian groups is presented in Table 2. There is a considerable range of allelic variability in African and Asian populations, although precise allele frequency estimates for specific racial subpopulations are only now beginning to emerge. The degree to which multiple copies of 2D6 genes are found on the 22nd chromosome varies in different ethnic populations. Specifically, North African and Mediterranean populations have a higher frequency of individuals who have multiple active copies of the 2D6 gene. In Ethiopia and Somalia, up to 30% of the population are ultra-rapid metabolizers (
4). For ultra-rapid metabolizers, an excess amount of 2D6 enzyme is produced, which results in the more rapid metabolism of 2D6 substrates. These patients require higher than the standard recommended dosage of 2D6 substrate medications to obtain a therapeutic blood level. A pharmacokinetic analysis of the variation in the clearance of nortriptyline in poor, intermediate, and ultra-rapid metabolizers has demonstrated the clinical importance of this phenotypic variation (
1).
The 2C19 gene
The 2C19 gene is located on the 10th chromosome and codes for the cytochrome P450 2C19 enzyme. The 2C19 gene is much less variable than the 2D6 gene. Only eight variants of the gene have been documented. The normal or “wild-type” allele is referred to as *1. All of the variant forms of the 2C19 gene do not produce any active enzyme.
Individuals with two copies of the *1 allele are considered normal and are referred to as extensive metabolizers. As with 2D6, an individual with one copy of the *1 allele and one inactive allele is usually classified as an intermediate metabolizer. Individuals with two deficient copies are always classified as poor metabolizers.
There are fewer 2C19 substrate psychotropic medications than there are 2D6 substrate psychotropic medications. Imipramine and amitriptyline should be used cautiously in patients who are poor metabolizers of C219. It has been suggested that poor 2C19 metabolizers be prescribed 60% of the standard dose of these tricyclic antidepressants (
3). Both citalopram and escitalopram are primarily metabolized by the 2C19 enzyme, although these drugs are also partially metabolized by 3A4 and to a lesser extent by 2D6. A dose adjustment to 60% of the standard dose of citalopram and escitalopram has been recommended for patients who are poor metabolizers of 2C19. Whereas sertraline is not predominantly metabolized by 2C19, it has been suggested that 75% of the usual recommended dose be prescribed for poor metabolizers of C219 (
3).
The SLC6A4 gene
The SLC6A4 gene is often referred to as the serotonin transporter gene. This gene is located on the long arm of the 17th chromosome and is also referred to as 5HTTR or SERT. An important variation of this gene is a 43-base deletion polymorphism located in the promoter region. This variation is regularly referred to as the “5-HTTLPR” polymorphism.
The “official” genetic abbreviation for the gene is SLC6A4. This designation indicates that the gene is a member of the solute carrier family of genes (i.e., the SLC family). The final number in the official designation of this gene indicates that SLC6A4 is the fourth member of the SLC gene family.
The SLC6A4 gene is 31 kilobases in size. This means that the gene is composed of a sequence of 31,000 nucleotide base pairs. Within the SLC6A4 gene, there are 14 exons or coding regions that define the sequences of messenger RNA, which ultimately determine the amino acid sequence that creates the serotonin transporter protein.
Studies of the SLC6A4 gene have traditionally classified subjects into three categories based on which of the two promoter region variations they have inherited. The first category refers to individuals who have two copies of the gene that both have the long form of the promoter region (i.e., long/long). The second category refers to those individuals who are homozygous for the short form of the promoter region (i.e., short/short). The short form is the result of a deletion of 43 base pairs. The third category refers to individuals who have one copy of the long form and one copy of the short form of the SLC6A4 gene (i.e., long/short). Individuals with the homozygous long form of the gene have been reported to have greater expression of the transporter molecule and therefore they have more rapid reuptake of the serotonin molecule in the presynaptic neuron. The long/long genotype has also been associated with a better response to selective serotonin reuptake inhibitors including fluvoxamine, paroxetine, and sertraline in Caucasian samples. This association has not been consistently demonstrated in Asian samples (
5,
6).
A number of studies have demonstrated that individuals with a homozygous short genotype are more likely to develop depression if they are subjected to stressful experiences (
7,
8). This finding has not been replicated in all studies, almost certainly in part because of the challenges in the quantitative measurement of “stress.”
The emergence of the promoter deletion polymorphism of the SLC6A4 gene is a relatively recent occurrence in the evolution of mammals. The long and short variations in the promoter region of the SLC6A4 gene occur only in primates (
9). The short form of the SLC6A4 gene has not been demonstrated in other mammals. Essentially, mice, dogs, and elephants do not have a short form of the SLC6A4 gene similar to the variant that is found in men and women. Promoter variations in the SLC6A4 gene do occur in other primates. However, the SLC6A4 gene has developed differently across primate species. Both the human long form and the human short form of the SLC6A4 gene appear to be unique to humans. Other primate species have evolved other species-specific variations that affect the transcription of the transporter molecule. Within
Homo sapiens, the evidence suggests that the long form is the original structure of the gene and that early in human development the short form evolved as a result of a deletion. Given the relatively high frequency of the short form in some ethnic populations, the hypothesis that the short form of SLC6A4 provides some adaptive advantage seems reasonable.
New variations of the SLC6A4 gene that influence the activity level of both the long and the short forms of the gene have been identified. One new variation is a single nucleotide polymorphism (SNP) that is located upstream to the portion of the promoter region with the 43-base deletion. The term “upstream” refers to the fact that the SNP is located toward the 5′ end of the gene. Two variants of this SNP have been referred to as the A form (i.e., adenine) and the G form (i.e., guanine). Individuals with the long form of the promoter region who also have the G version of this newly described SNP have been shown to have decreased SLC6A4 activity compared with that in individuals with the long form who have the A version of the SNP (
10).
The serotonin HTR2A gene
The serotonin 2A receptor gene (i.e., HTR2A) is located on the long arm of the 13th chromosome. The precise location of this gene is 13q14.13. The HTR2A gene is approximately 20 kilobases in length and consists of three exons and two introns. The 2A receptor is concentrated in the prefrontal-subcortical circuits of the neocortex.
A new polymorphism of the HTR2A gene has recently been described. The two forms of the polymorphism have been designated as the A (i.e., adenine) and G (i.e., guanine) variants. Patients with two copies of the HTR2A gene that contain the A variant of the polymorphism had an impressive 80% response rate to treatment with citalopram in an analysis of a large depression treatment effectiveness trial (
11). In contrast, only 62% of the patients with two copies of the HTR2A gene with the G variant had a positive response to citalopram. The allele frequency of this A polymorphism was 42% in patients who identified themselves as white and in only 6% of those who identified themselves as black. This sixfold difference in allele frequency of the A allele provides a plausible biological explanation for the clinical trial results, indicating that African American patients do not respond as well to selective serotonin reuptake inhibitors as Caucasian patients (
11).
This recently described polymorphism is located in the second intron of HTR2A. Because it is not located in a coding region, it is likely to influence the sensitivity of receptor through a regulatory effect on the expression of the serotonin 2A receptor gene.
Clinical implications
The clinical goal of genotyping both drug-metabolizing enzyme genes and target genes is to predict individual response to psychotropic medications. Clinicians can determine variations in the structure and function of a number of genes that produce enzymes involved in the metabolism of medication. It is also possible to genotype genes coding for receptors and transporter molecules that influence the therapeutic response of a patient to a specific psychotropic medication. Given the rapid development of the field of pharmacogenomics, it is safe to predict that the practice of psychopharmacology will become increasingly more evidence-based, and clinicians will be able to more accurately predict how individual patients will respond to specific medications.