Effect of CYP2D6 Genotype
We detected 11
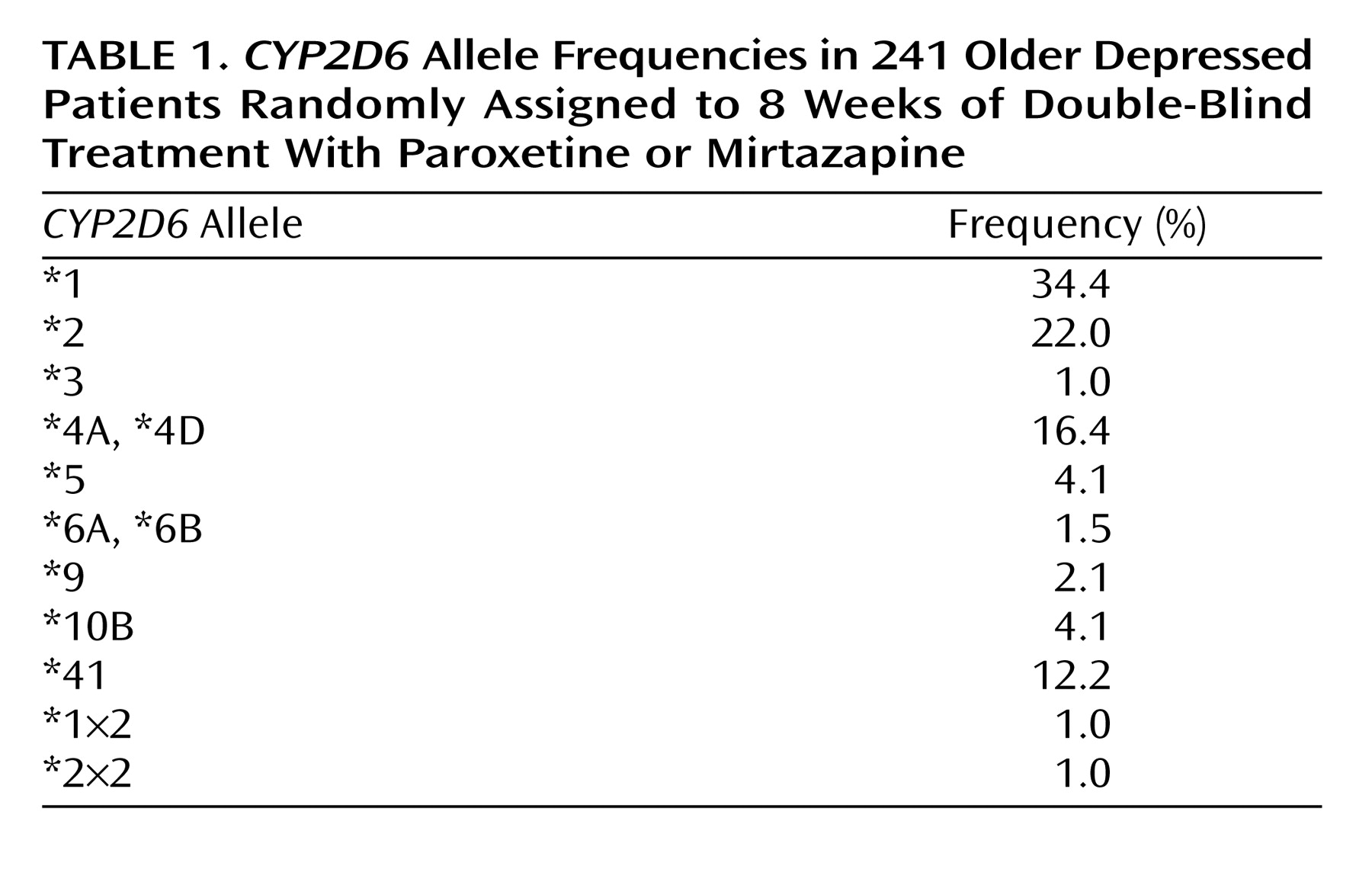
CYP2D6 alleles and 33 genotypes in 241 patients using oligonucleotide microarrays and additional assays for the *41 and *5 alleles and gene duplications.
CYP2D6 allele frequencies are given in
Table 1. Frequencies of the common
CYP2D6 alleles (*1, *2 and *41, *3, *4, *5) did not differ significantly (two-by-five chi-square test) from those reported for Caucasian populations
(28). The *10B allele was overrepresented in our sample, most likely because there were 15 ethnic minority patients included in the sample genotyped for
CYP2D6.
There were 42 patients (17.4%) with genotypes encoding poor (N=16) and intermediate metabolism (N=26). Ten patients (4.1%) carried gene duplications encoding ultrametabolism. There was no significant difference between the treatment groups in the frequencies of
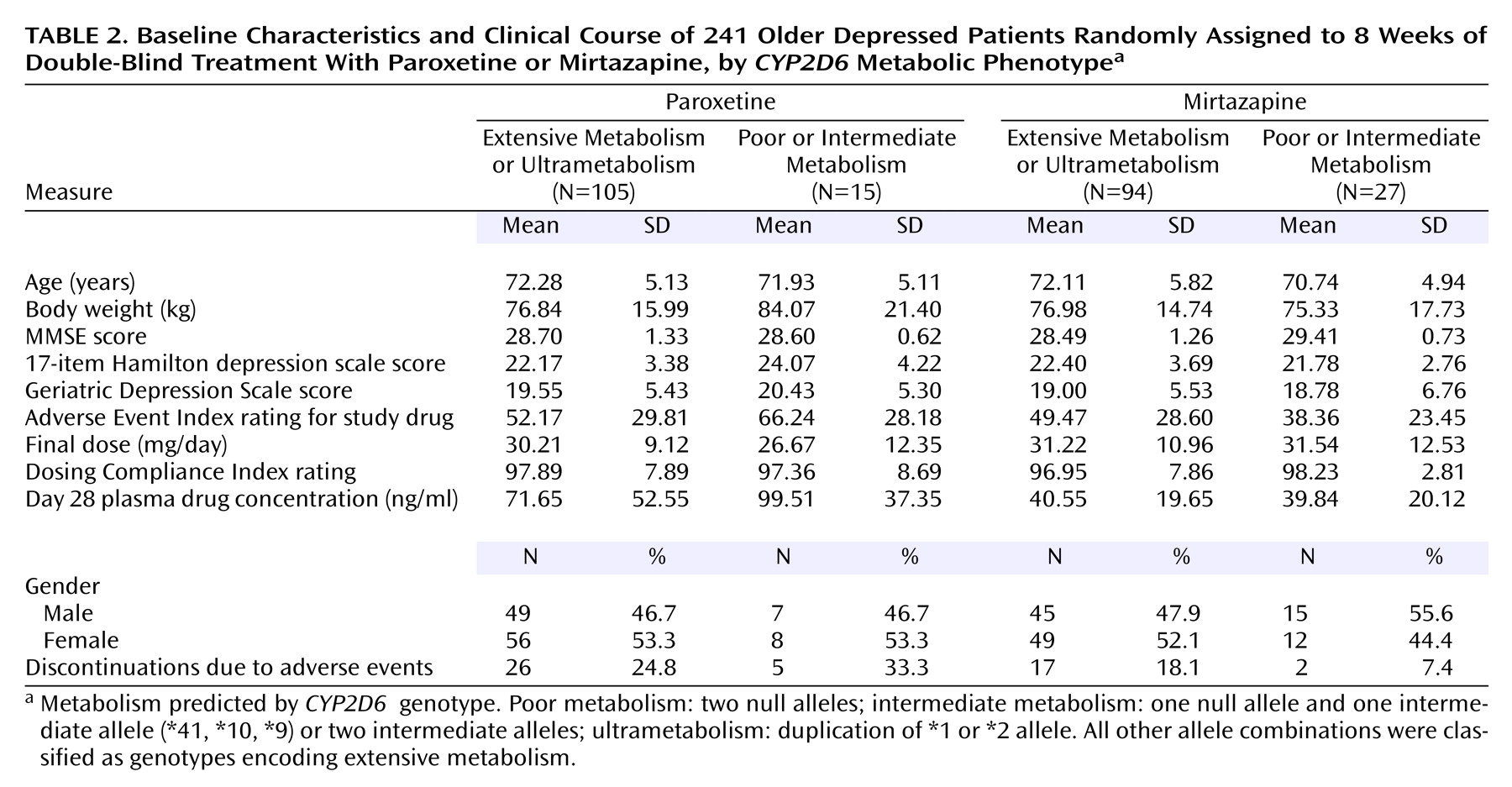
CYP2D6 genotype groups. Because of the small number of poor metabolizers and ultrametabolizers, analyses were performed on two combined groups: poor and intermediate metabolizers versus extensive metabolizers and ultrametabolizers. There were no significant differences between subjects classified as poor or intermediate metabolizers and subjects classified as extensive metabolizers or ultrametabolizers in age, gender distribution, ethnicity, baseline body weight, baseline MMSE, or baseline depression rating scale scores (
Table 2). For both medications, patients with genotypes predicting poor or intermediate metabolism showed no differences in the severity of adverse events or the frequency of discontinuations from those with genotypes encoding extensive and ultrametabolism. There were no differences between groups in final daily dose achieved or in dosing compliance. Plasma drug levels obtained after 4 weeks of treatment showed no significant differences for either drug between patients with predicted poor or intermediate metabolism and others.
CYP2D6 genotype had no effect on depression measures for either drug. Reanalysis of data with patients carrying one null allele and a functional allele (N=64) classified as intermediate metabolizers yielded identical results. Results were similar when data for the 222 Caucasian patients with
CYP2D6 genotypes were analyzed alone.
An analysis of variance showed no significant interaction between concurrent medication (debrisoquine hydroxylase inhibitor or substrate) and CYP2D6 genotype effects on severity of adverse events. There was no interaction between concurrent medication and study medication and no significant three-way interaction. These results indicate that concurrent medication did not interact with CYP2D6 genotype or study medication to affect the severity of adverse events.
Effect of HTR2A Genotype
In the full sample,
HTR2A 102 T/C allele frequencies were C=0.575, T=0.425. Genotype frequencies did not differ significantly from Hardy-Weinberg equilibrium. We analyzed clinical results by comparing patients with the C/C genotype with others because this dichotomy was shown to predict outcome in patients treated with clozapine
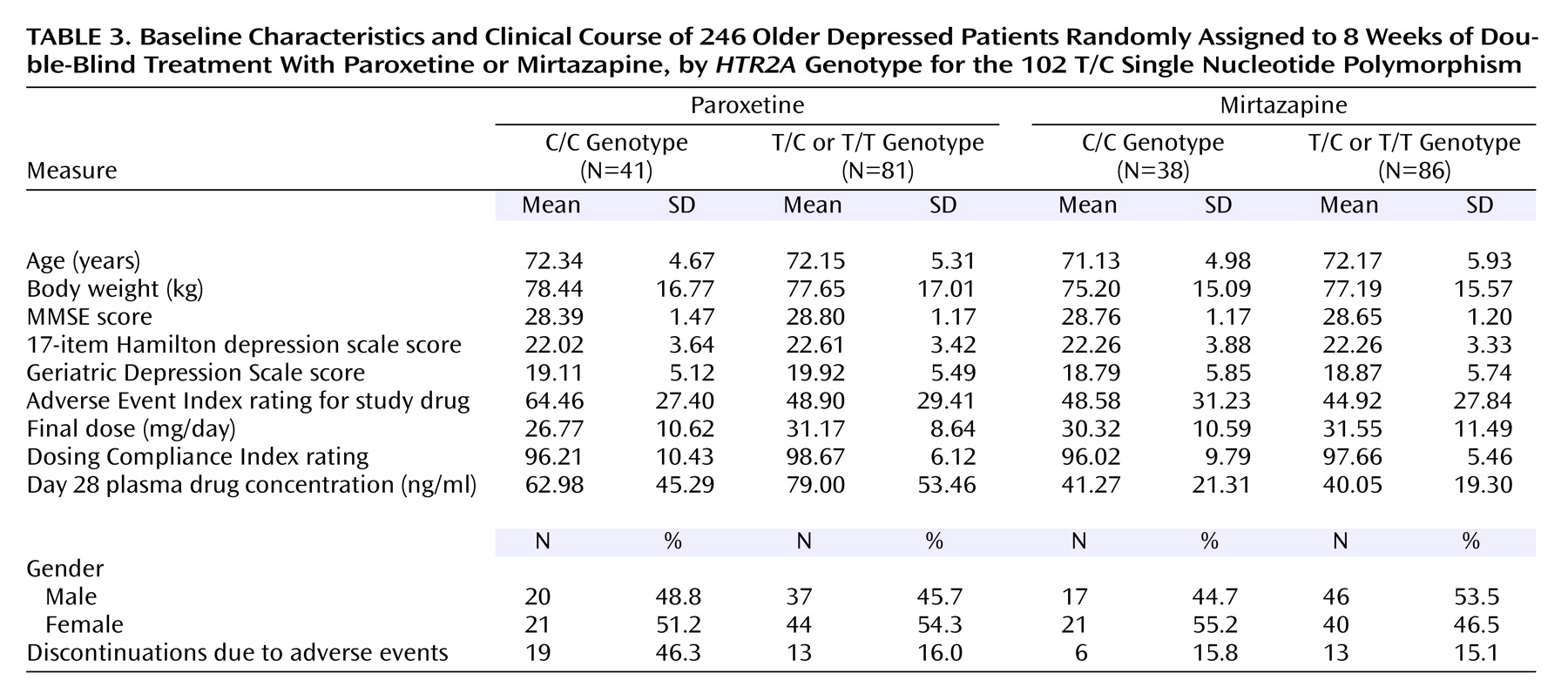
(15). Of paroxetine-treated patients, 41 (33.6%) had the C/C genotype, whereas 81 (66.4%) were T/C or T/T. For mirtazapine, there were 38 patients (30.6%) with the C/C genotype, whereas 86 (69.4%) were T/C or T/T. These frequencies were not significantly different between the paroxetine and mirtazapine treatment groups. There were no significant differences between patients with the C/C genotype and others in age, gender distribution, ethnicity, baseline body weight, plasma drug concentrations, baseline cognition, or severity of depression at baseline for either treatment group (
Table 3).
Unlike
CYP2D6 variation, the
HTR2A 102 T/C SNP had a major effect on paroxetine side effects and discontinuations. There were significantly more discontinuations due to adverse events for C/C paroxetine-treated patients (46.3%) than for those with the T/C and T/T genotypes (16%) (Cochran-Mantel-Haenszel test: χ
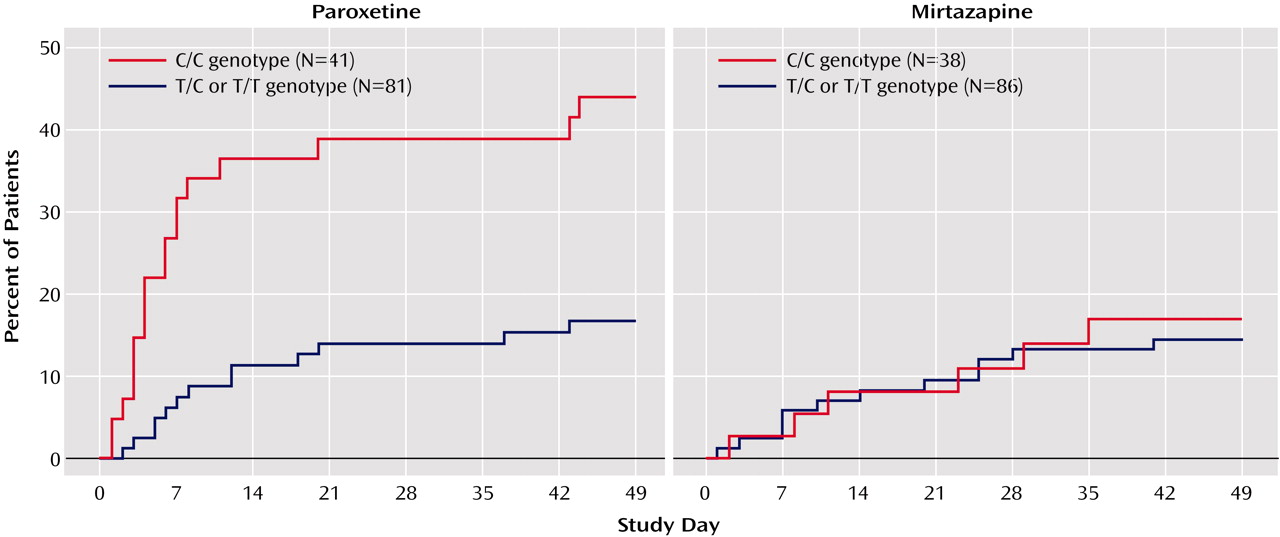
2=12.8, df=1, p=0.001). Survival analyses showed that paroxetine-treated patients with the C/C genotype were significantly more likely to discontinue treatment because of adverse events than were other patients at all assessment points (p=0.001 for all points, log rank chi-square tests) (
Figure 1). The severity of side effects in paroxetine-treated patients with the C/C genotype was also greater (F=4.61, df=1, 179, p=0.03). During the first week of treatment, C/C patients took significantly less paroxetine than others (F=7.64, df=1, 108, p=0.007), indicating these patients did not comply with medication instructions. Paroxetine-treated patients discontinued early because of gastrointestinal complaints (vomiting, nausea, diarrhea) (N=7), somnolence and difficulty concentrating (N=7), agitation and sleep disturbance (N=6), and other side effects (dizziness, sweating, headache, sexual dysfunction) (N=7).
In contrast, among mirtazapine-treated patients, there were no differences between C/C patients and other subjects in severity of adverse events, final daily dose, dosing compliance, plasma levels, early discontinuations, or dropouts due to adverse events. Survival analysis showed no difference between those with the C/C genotype and others in discontinuations at any point during the study (
Figure 1).
We also performed three-level analyses comparing patients with C/C, T/C, and T/T genotypes. For the full sample, genotype frequencies were as follows: C/C=32.1%, T/C=50.8%, and T/T=17.1%. There was no significant difference between the treatment groups in the distribution of HTR2A genotypes (paroxetine group: C/C=33.6%, T/C=51.6%, T/T=14.8%; mirtazapine group: C/C=30.6%, T/C=50.0%, T/T=19.4%). There were no significant differences among the genotype groups in age, gender distribution, ethnicity, body weight, or baseline clinical measures for either treatment group. Survival analyses showed that among paroxetine-treated patients, there was a positive linear relationship between the number of C alleles and the probability of discontinuation because of adverse events at all assessment points (p=0.025–0.009, log rank chi-square tests). No effect of HTR2A genotype was seen among mirtazapine-treated patients.
Although there were no significant differences between HTR2A genotype groups in the frequency of ethnic minorities, to minimize error due to population stratification data for Caucasians (N=117 for mirtazapine, N=109 for paroxetine) were analyzed separately. There were no significant differences between C/C carriers and other Caucasian patients in baseline demographics and clinical characteristics for either treatment group. For paroxetine-treated patients, survival analyses showed that C/C patients had a significantly greater probability of discontinuation due to adverse events at every assessment point in comparison with other patients (p<0.005 for all points, log rank chi-square tests), whereas among mirtazapine-treated Caucasian patients there was no difference.
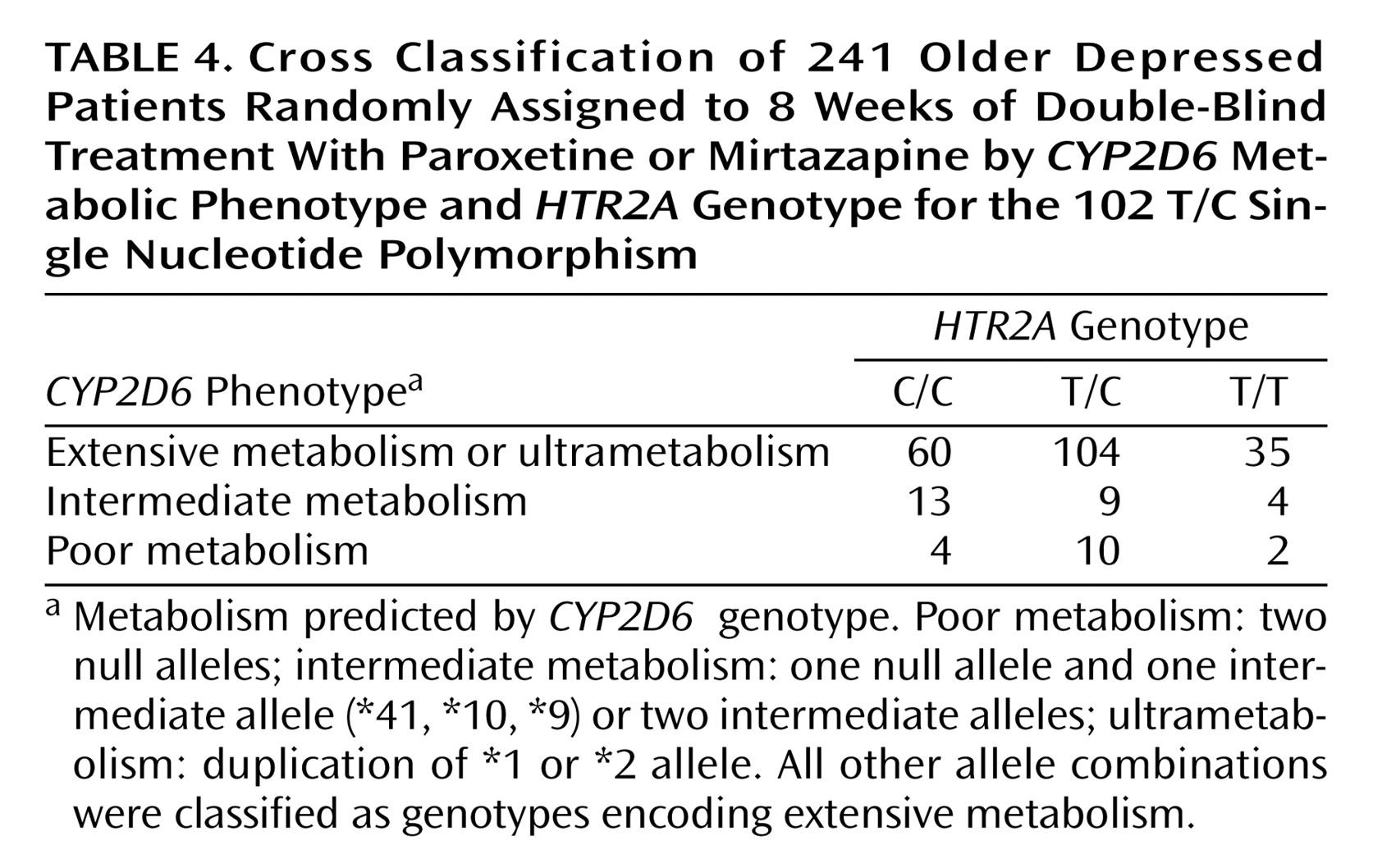
Chi-square tests showed no association between
HTR2A 102 T/C genotype and
CYP2D6 genotype for the full cohort (
Table 4), as well as when the paroxetine and mirtazapine cohorts were considered separately. This means it is unlikely that effects observed for
HTR2A C/C genotype were due to an association with a particular
CYP2D6 genotype category.