Identification of Two Heritable Cross-Disorder Endophenotypes for Tourette Syndrome
Abstract
Objective:
Method:
Results:
Conclusions:
Method
Samples
Procedure
Statistical Analyses
Exploratory factor analyses.
Latent class analyses.
Replication sample analyses.
Heritability estimates.
Polygenic burden analyses.
Results
Sample Characteristics
| Original Sample | Validation Sample | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Characteristic | Probands Only (N=1,191) | Probands and Family Members (N=3,494) | Probands Only (N=527) | Probands and Family Members (N=882) | ||||||||
| N | Mean | SD | N | Mean | SD | N | Mean | SD | N | Mean | SD | |
| Age (years) | 1,191 | 15.3 | 10.0 | 3,494 | 30.6 | 17.2 | 522 | 21.4 | 15.6 | 864 | 30.0 | 17.5 |
| Tourette syndrome | ||||||||||||
| Age at onset (years) | 1,131 | 5.8 | 2.5 | 1,692 | 6.1 | 2.7 | 521 | 6.2 | 2.8 | 569 | 6.3 | 2.8 |
| Severitya | 1,181 | 11.4 | 2.6 | 3,490 | 4.1 | 4.6 | 478 | 11.7 | 2.5 | 764 | 7.9 | 5.6 |
| OCD | ||||||||||||
| Age at onset (years) | 442 | 7.1 | 4.0 | 712 | 8.2 | 5.3 | 249 | 7.4 | 3.8 | 299 | 8.0 | 4.4 |
| Severitya | 694 | 4.3 | 3.4 | 2,442 | 3.0 | 3.3 | 450 | 5.0 | 3.9 | 681 | 3.9 | 3.8 |
| Total N | N | % | Total N | N | % | Total N | N | % | Total N | N | % | |
| Male | 1,191 | 944 | 79.3 | 3,494 | 2,140 | 61.2 | 527 | 408 | 77.4 | 882 | 581 | 65.9 |
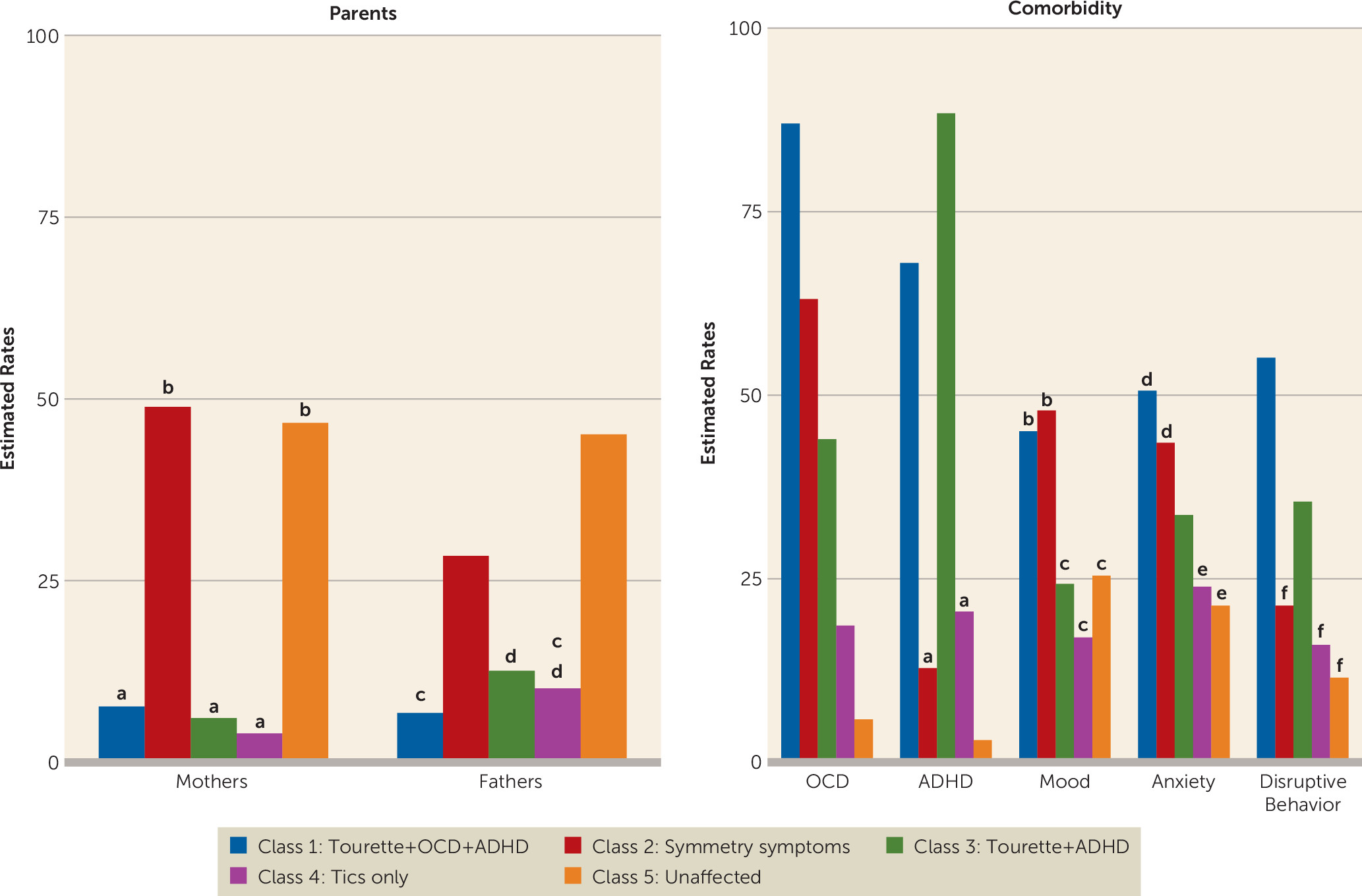
| Parental history of Tourette syndrome or chronic motor or vocal tic disorder | 864 | 385 | 44.6 | — | — | — | — | — | — | — | — | — |
| Personal history | ||||||||||||
| OCD | 1,135 | 570 | 50.2 | 3,286 | 1,125 | 34.2 | 504 | 219 | 43.5 | 847 | 336 | 39.7 |
| ADHD | 1,116 | 628 | 56.3 | 3,220 | 1,013 | 31.5 | 494 | 181 | 36.6 | 830 | 293 | 35.3 |
| Mood disorders | 498 | 132 | 26.5 | 1,603 | 487 | 30.4 | — | — | — | — | — | — |
| Anxiety disorders | 507 | 176 | 34.7 | 1,620 | 515 | 31.8 | — | — | — | — | — | — |
| Disruptive behavior disorders | 390 | 121 | 31.0 | 662 | 192 | 29.0 | — | — | — | — | — | — |
Exploratory Factor Analyses
| Factor Loading | |||||
|---|---|---|---|---|---|
| Category | Item | Factor 1: Tics (Cronbach’s alpha=0.88) | Factor 2: Obsessive-Compulsive Symptoms (Cronbach’s alpha=0.92) | Factor 3: Disinhibition (Cronbach’s alpha=0.77) | Factor 4: Symmetry Symptoms (Cronbach’s alpha=0.87) |
| Tourette | Lift chin | 0.67 | 0.02 | –0.09 | –0.06 |
| Tourette | Knee-bending | 0.66 | 0.02 | 0.07 | 0.14 |
| Tourette | Teeth bearing | 0.63 | 0.10 | 0.07 | –0.17 |
| Tourette | Touch chin to shoulder | 0.60 | 0.04 | –0.08 | –0.08 |
| Tourette | Tensing buttocks | 0.58 | 0.02 | –0.16 | 0.18 |
| Tourette | Deep knee bending | 0.57 | –0.14 | 0.19 | 0.28 |
| Tourette | Quick eye turn | 0.56 | 0.13 | –0.07 | –0.04 |
| Tourette | Tensing abdomen | 0.56 | –0.03 | –0.15 | 0.12 |
| Tourette | Lip pouting | 0.56 | 0.13 | –0.04 | –0.11 |
| Tourette | Rolling eyes to one side | 0.55 | 0.02 | –0.10 | –0.01 |
| Tourette | Broadening nostrils | 0.55 | 0.20 | –0.22 | –0.08 |
| Tourette | Smiling | 0.54 | 0.07 | 0.03 | –0.04 |
| Tourette | Opening eyes wide | 0.52 | 0.10 | –0.10 | 0.01 |
| Tourette | Flexing or extending ankle | 0.52 | 0.12 | –0.13 | 0.07 |
| Tourette | Kicking | 0.52 | –0.01 | 0.32 | 0.03 |
| Tourette | Sticking tongue out | 0.51 | 0.06 | 0.24 | –0.13 |
| Tourette | Shrugging shoulders | 0.51 | 0.00 | 0.00 | –0.07 |
| Tourette | Nose twitching | 0.50 | 0.06 | –0.18 | –0.06 |
| Tourette | Squatting | 0.49 | –0.17 | 0.19 | 0.29 |
| Tourette | Biting tongue | 0.48 | 0.13 | –0.03 | 0.02 |
| Tourette | Touching | 0.48 | –0.14 | 0.22 | 0.35 |
| Tourette | Squinting | 0.48 | 0.13 | –0.15 | –0.01 |
| Tourette | Tapping | 0.48 | –0.07 | 0.12 | 0.26 |
| Tourette | Jerking shoulder | 0.47 | 0.00 | –0.05 | 0.02 |
| Tourette | Counting with fingers | 0.46 | 0.02 | 0.03 | 0.20 |
| Tourette | Bending or gyrating | 0.45 | –0.03 | 0.31 | 0.07 |
| Tourette | Pulling back pencil | 0.43 | –0.02 | 0.13 | 0.17 |
| Tourette | Throwing head back | 0.43 | 0.00 | –0.09 | 0.04 |
| Tourette | Skipping | 0.42 | –0.08 | 0.28 | 0.05 |
| Tourette | Chewing on lip | 0.42 | 0.15 | –0.04 | –0.11 |
| Tourette | Unusual postures | 0.42 | –0.03 | 0.28 | 0.09 |
| Tourette | Slower movements | 0.41 | –0.02 | 0.20 | 0.41 |
| OCD | Needs to touch, tap, or rub things | 0.41 | 0.13 | 0.14 | 0.39 |
| Tourette | Eye blinking | 0.41 | 0.15 | –0.27 | –0.04 |
| Tourette | Syllables | 0.40 | –0.16 | 0.40 | 0.05 |
| OCD | Contamination concern | 0.00 | 0.77 | –0.23 | –0.05 |
| OCD | Hoarding obsessions | –0.24 | 0.76 | 0.43 | –0.01 |
| OCD | Illness concern | 0.04 | 0.73 | –0.13 | 0.02 |
| OCD | Hoarding compulsions | –0.21 | 0.70 | 0.42 | –0.03 |
| OCD | Dirt or germ obsessions | –0.04 | 0.70 | –0.16 | 0.08 |
| OCD | Fears harming others if not careful | 0.03 | 0.66 | 0.14 | 0.07 |
| OCD | Fears responsible for something terrible | 0.03 | 0.65 | 0.05 | 0.19 |
| OCD | Thoughts may influence the outcome of some events if does certain things | –0.02 | 0.63 | –0.09 | 0.32 |
| OCD | Checks no self-harm | 0.03 | 0.62 | 0.08 | –0.08 |
| OCD | Fears harming others | 0.08 | 0.62 | 0.01 | 0.09 |
| OCD | Fears losing things | –0.02 | 0.62 | 0.10 | 0.18 |
| OCD | Contamination compulsions | 0.08 | 0.61 | –0.12 | 0.04 |
| OCD | Contamination obsessions | 0.22 | 0.61 | –0.08 | 0.03 |
| OCD | Checks nothing terrible | 0.05 | 0.60 | 0.11 | –0.02 |
| OCD | Fears will steal things | 0.06 | 0.60 | 0.22 | –0.16 |
| OCD | Religious obsessions | 0.06 | 0.60 | 0.03 | 0.06 |
| OCD | Concerned with bodily waste | 0.05 | 0.59 | –0.03 | 0.09 |
| OCD | Morality obsessions | 0.03 | 0.58 | –0.08 | 0.20 |
| OCD | Compulsions to prevent harm | 0.25 | 0.57 | –0.03 | 0.10 |
| OCD | Urges to offend | –0.05 | 0.57 | 0.56 | –0.10 |
| OCD | Checks no harm | 0.07 | 0.57 | 0.08 | 0.12 |
| OCD | Violent obsessions | 0.02 | 0.56 | 0.13 | 0.08 |
| OCD | Fears self-harm | 0.19 | 0.56 | –0.03 | 0.04 |
| OCD | Thoughts can influence the outcome of some events if does certain things | –0.02 | 0.54 | –0.10 | 0.33 |
| OCD | Mental rituals | –0.04 | 0.53 | 0.00 | 0.34 |
| OCD | Fears acting on an unwanted impulse | 0.09 | 0.53 | 0.29 | 0.12 |
| OCD | Self-harm urges | 0.11 | 0.52 | 0.35 | 0.01 |
| OCD | Superstitious fears | 0.02 | 0.50 | –0.14 | 0.35 |
| OCD | Sexual obsessions | 0.13 | 0.49 | 0.18 | 0.14 |
| OCD | Need to confess | 0.00 | 0.49 | 0.12 | 0.18 |
| OCD | Animal obsessions | –0.08 | 0.48 | 0.00 | 0.12 |
| OCD | Need to explore surroundings | –0.14 | 0.46 | 0.42 | 0.03 |
| OCD | Concerned with a part of body or appearance | 0.02 | 0.45 | 0.05 | 0.20 |
| OCD | Superstitious behaviors | 0.03 | 0.42 | –0.08 | 0.34 |
| OCD | Urges to do sudden and reckless things | 0.11 | 0.40 | 0.30 | 0.13 |
| Tourette | Coprolalia | 0.25 | 0.05 | 0.71 | 0.00 |
| Tourette | Copropraxia | 0.27 | 0.06 | 0.66 | –0.08 |
| Tourette | Echolalia | 0.25 | –0.10 | 0.56 | 0.23 |
| OCD | Urges to be destructive | 0.02 | 0.51 | 0.55 | –0.02 |
| OCD | Urges to injure or mutilate others | –0.10 | 0.52 | 0.55 | –0.17 |
| Tourette | Words | 0.31 | 0.04 | 0.54 | 0.05 |
| Tourette | Palilalia | 0.27 | –0.02 | 0.50 | 0.25 |
| Tourette | Animal or bird noises | 0.26 | 0.06 | 0.41 | –0.11 |
| OCD | Symmetry obsessions | –0.07 | 0.09 | –0.04 | 0.82 |
| OCD | Needs certain things to be symmetrical | –0.02 | 0.06 | –0.01 | 0.76 |
| OCD | Ordering or arranging compulsions | –0.10 | 0.11 | 0.07 | 0.73 |
| OCD | Needs to have certain things evened up | 0.08 | 0.06 | –0.05 | 0.73 |
| OCD | Thoughts about evening things up | 0.07 | 0.08 | –0.01 | 0.73 |
| OCD | Thoughts about lining things up | –0.01 | 0.09 | –0.01 | 0.71 |
| OCD | Obsessions about exactness | –0.03 | 0.20 | –0.05 | 0.70 |
| OCD | Rereads or rewrites things | 0.06 | 0.20 | 0.05 | 0.59 |
| OCD | Needs to repeat routine activities | 0.03 | 0.14 | 0.11 | 0.58 |
| OCD | Counting compulsions | 0.11 | 0.14 | 0.05 | 0.54 |
| OCD | Has to do things the same way every time | –0.11 | 0.22 | 0.21 | 0.47 |
| OCD | Needs to know or remember certain things | 0.08 | 0.36 | 0.05 | 0.44 |
| OCD | Checks that did not make mistakes | 0.05 | 0.40 | –0.09 | 0.43 |
| Tourette | Writing the same letter or word over and over | 0.20 | 0.04 | 0.03 | 0.42 |
| OCD | Checks things | 0.11 | 0.35 | –0.11 | 0.42 |
Latent Class Analyses
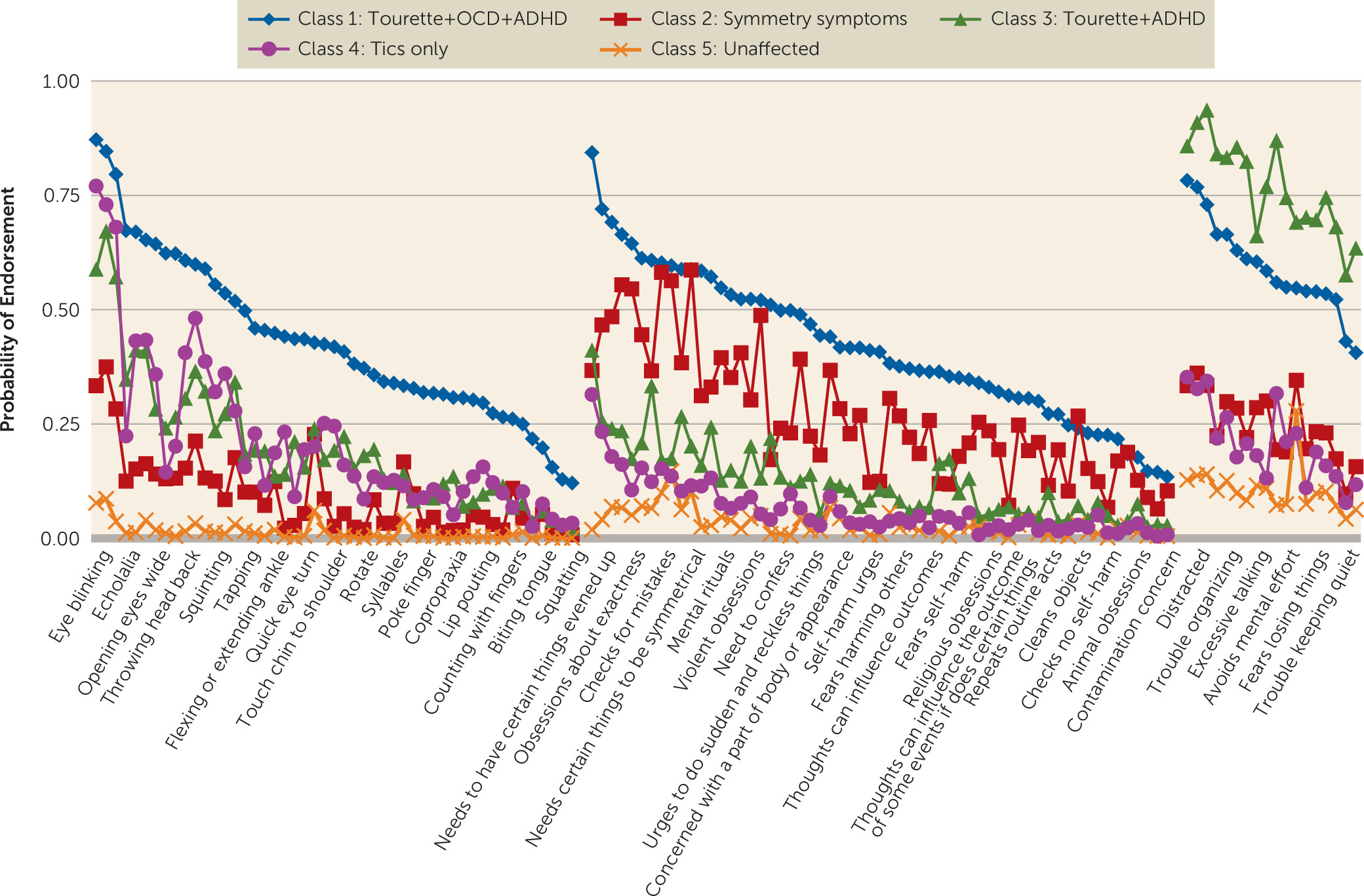
Replication Analyses
Heritability Analyses
| Type of Analysis | H2r | SE | p |
|---|---|---|---|
| Latent class analyses | |||
| Class 1: Tourette+OCD+ADHD | 0.63 | 0.09 | 2.0×10–13 |
| Class 2: Symmetry symptoms | 0.38 | 0.10 | 0.001 |
| Class 3: Tourette+ADHD | 0.47 | 0.08 | 3.3×10–29 |
| Class 4: Tics only | 0.28 | 0.08 | 0.001 |
| Class 5: Unaffected | — | — | — |
| Exploratory factor analyses | |||
| Factor 1: Tics | 0.25 | 0.04 | 2.6×10–13 |
| Factor 2: Obsessive-compulsive symptoms | 0.46 | 0.03 | 8.6×10–41 |
| Factor 3: Disinhibition | 0.35 | 0.03 | 4.2×10–34 |
| Factor 4: Symmetry symptoms | 0.39 | 0.03 | 7.2×10–31 |
Polygenic Burden
Discussion
Limitations
Acknowledgments
Footnotes
Supplementary Material
- View/Download
- 721.85 KB
References
Information & Authors
Information
Published In
History
Keywords
Authors
Funding Information
Metrics & Citations
Metrics
Citations
Export Citations
If you have the appropriate software installed, you can download article citation data to the citation manager of your choice. Simply select your manager software from the list below and click Download.
For more information or tips please see 'Downloading to a citation manager' in the Help menu.
View Options
View options
PDF/EPUB
View PDF/EPUBLogin options
Already a subscriber? Access your subscription through your login credentials or your institution for full access to this article.
Personal login Institutional Login Open Athens loginNot a subscriber?
PsychiatryOnline subscription options offer access to the DSM-5-TR® library, books, journals, CME, and patient resources. This all-in-one virtual library provides psychiatrists and mental health professionals with key resources for diagnosis, treatment, research, and professional development.
Need more help? PsychiatryOnline Customer Service may be reached by emailing [email protected] or by calling 800-368-5777 (in the U.S.) or 703-907-7322 (outside the U.S.).

