Depression is a leading cause of disability burden globally (
1). Depression is a particular concern among older adults, who are a growing population segment, and in whom depression may contribute to mobility limitation leading to disability and premature mortality (
2). Depression in older adults shares etiological features with depression in younger adults, reflecting a combination of genetic and environmental influences (
3). A critical question is why events that can trigger depressive episodes may affect some individuals differently than others.
Experience of stressful life events is associated with risk of depression (
4,
5). Yet not all stress-exposed individuals become depressed. A controversial hypothesis is that genetic differences between individuals modify the influence of life-stress on depression (
6). This “diathesis-stress” hypothesis is supported by family-based genetic studies that find individuals with familial liability to depression are more vulnerable to developing depression following stress exposure (
7). Molecular genetic evidence for the diathesis-stress hypothesis from candidate-gene studies is contested, partly because of concerns about the candidate-gene approach (
8). Genome-wide association studies (GWAS) offer opportunities to move beyond arguments about candidate genes. GWAS discoveries in depression have yielded mixed results in tests of diathesis-stress models (
9–
11). A recent GWAS of subjective well-being (
12) offers the opportunity to evaluate the opposite end of the continuum of genetic liability.
Subjective well-being represents a dimension of so-called “positive psychology” and may reflect the opposite end of an affective continuum from depression (
13). Like depression, subjective well-being is heritable, with a portion of this heritability reflecting genetic influences shared with depression (
14). Theory predicts increased subjective well-being should buffer against deleterious consequences of stressful life events (
15).
We conducted a polygenic score study to test whether genetic predisposition to greater subjective well-being buffered against risk of depression following experience of a stressful life event: the death of one’s spouse. We studied depressive symptoms in a cohort of older adults and their spouses followed longitudinally as part of the U.S. Health and Retirement Study (HRS). We combined whole-genome single-nucleotide polymorphism (SNP) data already generated for the cohort and results from GWAS of subjective well-being in over 100,000 adults by the Social Science Genetic Association Consortium (
12) to calculate cohort members’ subjective well-being polygenic scores. Analysis tested whether higher subjective well-being polygenic scores buffered survivors’ risk of developing depressive symptoms following the death of their spouse. For comparative purposes, we repeated the analysis using polygenic scores based on GWAS of major depressive disorder by the Psychiatric Genomics Consortium (
16) and GWAS of depressive symptoms by the Social Science Genetic Association Consortium (
12).
Results
Higher Well-Being Polygenic Score Was Associated With Reduced Depressive Symptoms
HRS respondents with higher polygenic scores experienced fewer depressive symptoms during follow-up (b=−0.10, confidence interval [CI]=−0.13 to –0.07, p<0.001) (
Figure 1; also see Table S1/Model 1 in the
online data supplement). This effect size is consistent with previous analysis of the well-being polygenic score and depression (
12). The genetic association with depressive symptoms was similar for men and women (p=0.803 for test of interaction between polygenic score and sex).
Well-Being Polygenic Score Was Not Associated With Likelihood of Experiencing Death of a Spouse
Genetic liability to depression may influence exposure to stressful life events in older adults. Therefore, before testing the genetic buffering hypothesis, we evaluated possible gene-environment correlation between subjective well-being polygenic score and likelihood of experiencing spousal death. We found no evidence of such gene-environment correlation; HRS respondents’ polygenic scores were not related to their likelihood of experiencing the death of their spouse (odds ratio=0.97, CI=0.92–1.02, p=0.219).
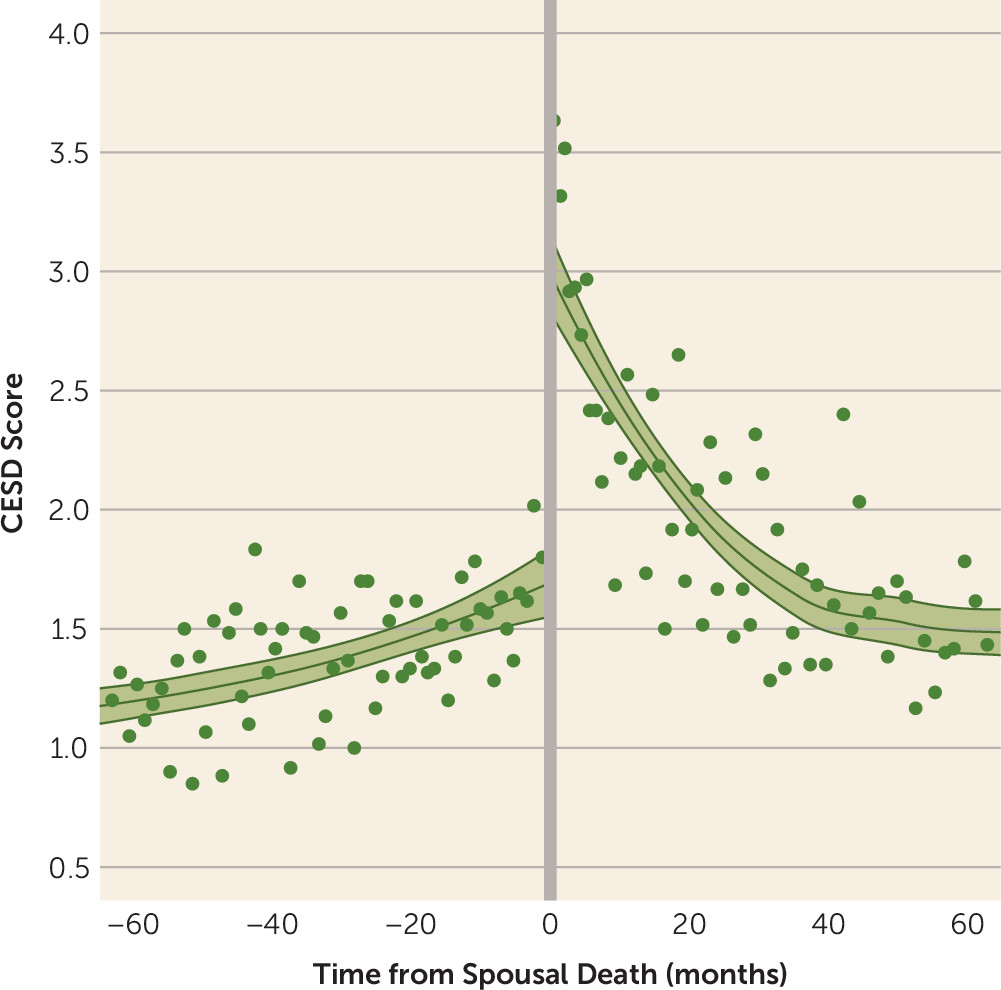
Death of a Spouse Was Associated With an Immediate Increase in Depressive Symptoms, Followed by a Gradual Return Toward Baseline Over the Following Years
HRS respondents who experienced death of their spouse experienced a discontinuity in depressive symptoms around the time of the death. In local regression analysis, respondents’ depressive symptoms trended slightly upwards in the months immediately preceding the death, rose sharply at the time of death, and gradually returned toward baseline over the following two years (
Figure 2). This pattern is consistent with previous reports on the course of depressive symptoms around spousal death (
28).
Nonlinear regression analysis estimated the increase in Center for Epidemiological Studies Depression score associated with spousal death to be 1.97 points (95% CI 1.5–2.4, p≤0.001). This increase was smaller for respondents interviewed farther from the time of their spouse’s death. Based on trajectories estimated from the model, the increase in depressive symptoms that occurred following the death of a spouse attenuated by 12 months after the death, with modest further attenuation through 24 months (see Table S1/Model 2 in the online data supplement).
Genetics of Subjective Well-Being Buffered Against Increase in Depressive Symptoms Following Death of a Spouse
HRS respondents with higher polygenic scores experienced flatter trajectories of depressive symptoms around the time of their spouse’s death compared with peers who also lost their spouse and had lower polygenic scores. In local regression analysis, HRS respondents with higher well-being polygenic scores experienced fewer depressive symptoms in the months leading up to the death and a smaller increase in depressive symptoms following the death compared with other survivors who had lower polygenic scores (
Figure 3).
In nonlinear regression analysis, each standard deviation increase in polygenic score buffered the increase in depressive symptoms following spousal death by 0.60 Center for Epidemiological Studies Depression points (95% CI=0.19–1.02, p=0.004). In context, this effect estimate suggests that a person with a polygenic score one standard deviation above the population mean should experience about half as much increase in depressive symptoms following death of a spouse compared with a peer whose polygenic score was one standard deviation below the population mean. Respondents’ polygenic scores were not related to the rate of attenuation in depressive symptoms following the death (γ=0.33, p=0.739 [also see Table S1 in the online data supplement]).
Sensitivity Analyses
We conducted sensitivity analyses to evaluate consistency of findings. First, we repeated the analysis including all HRS respondents who self-reported as being of non-Hispanic white race/ethnicity (N=9,453 [N=1,829 who survived the death of a spouse]). Findings were similar to those in the analysis sample determined by genetically defined ancestry (see Table S2 in the data supplement).
Second, we repeated the analysis using polygenic scores derived from two published GWAS of depression, the Psychiatric Genomics Consortium’s GWAS of major depressive disorder (
16) and the Social Science Genetic Association Consortium GWAS of depressive symptoms (
12). We analyzed genetic overlap among phenotypes using linkage disequilibrium score regression (
29) and polygenic score methods. Linkage disequilibrium score regression estimates genetic correlations (rG) from GWAS summary statistics. We implemented linkage disequilibrium score regression using the LDSC Python software package (
https://github.com/bulik/ldsc) following the method described by the Social Science Genetic Association Consortium (
12). Previous linkage disequilibrium score regression analysis estimated rG between subjective well-being and depressive symptoms to be r=−0.80 (
12). Genetic correlation computed from subjective well-being GWAS results excluding the HRS sample was similar (for depressive symptoms: rG=−0.81, SE=0.06; for major depressive disorder: rG=−0.78, SE=0.12). Empirical correlations between polygenic scores were lower (|r|=0.15–0.32). All genetic correlations and polygenic score correlations were statistically significant at a p value <0.001.
Results from analysis of depression polygenic scores paralleled results from analysis of the subjective well-being polygenic score. HRS respondents with higher depression polygenic scores had higher levels of depressive symptoms across HRS follow-up. Among those who survived the death of a spouse, higher depression polygenic scores predicted a greater increase in depressive symptoms following the death (
Figure 4). In nonlinear regression analysis, each standard deviation increase in the depressive symptoms polygenic score amplified the increase in depressive symptoms following spousal death by 0.69 Center for Epidemiological Studies Depression points (95% CI=0.24–1.14, p=0.003). The result was similar for the major depressive disorder polygenic score (b=0.63, 95% CI=0.21–1.05, p=0.003). Results from analysis of depression polygenic scores are reported in Table S3 in the
online data supplement.
Discussion
We tested whether genetics discovered in GWAS of subjective well-being buffered against development of depressive symptoms following death of a spouse. We analyzed data on depressive symptoms in 8,588 individuals followed longitudinally as part of the HRS. During follow-up, 1,647 of these individuals experienced the death of a spouse. HRS respondents with higher polygenic scores experienced fewer depressive symptoms during follow-up. For those whose spouse died during the follow-up period, having a higher polygenic score buffered against increased depressive symptoms following the death. Having a low polygenic score for depression had similar buffering effects on depression risk as having a high polygenic score for subjective well-being. Magnitudes of genetic effects were small.
This evidence of genetic buffering against depression following a stressful life event provides proof of concept for further investigation of the genetics of subjective well-being as a modifier of the life-stress to depression link. Studies of gene-environment interplay are controversial (
30). Statistical and theoretical models are contested. The precise nature of any “interaction” between the stress of losing a spouse and well-being-related genetic background remains uncertain. What we find is that individuals who carry more well-being-associated alleles tend to develop fewer additional depressive symptoms following the death of a spouse. This “buffering” is independent of the “main effect” of genetics in the absence of a spousal death. Buffering is also not affected by any “gene-environment correlation” between the genetics of subjective well-being and exposure to death of a spouse; HRS respondents’ subjective well-being polygenic scores were unrelated to their risk of losing a spouse during follow-up. Finally, we detected consistent evidence of buffering using two different statistical specifications, local regression and nonlinear regression, which take into account time elapsed between the stressful event and the measurement of depressive symptoms.
Our findings have implications for relevance of well-being genetics in clinical approaches to bereavement and for research into gene-environment interplay. Clinically, genetic testing based on current knowledge is unlikely to be useful in planning for or managing bereavement. Effect sizes in our study were too small to be useful for predicting outcomes of individual patients. It is possible that larger-scale GWAS will furnish more predictive polygenic scores with greater clinical relevance. However, even with very large GWAS, the predictive power of polygenic scores for common, complex traits will be limited (
31). Instead, our results may inform clinical research into candidate intervention targets to promote resilience in bereavement and in the context of other stressful experiences. Specifically, our findings raise questions about what behaviors or psychological processes mediate buffering effects. Do individuals with higher well-being polygenic scores process or cope with stress differently (
32)? Do they cultivate different social support networks or interact with them in different ways? Studies to identify such mediators of genetic associations can inform interventions designed to promote resilience. Finally, given that we observed essentially the same result in our analysis of polygenic scores for subjective well-being and depression, research is needed to evaluate whether behavioral and psychological mediators of genetic influences measured by these scores are the same or different.
High genetic correlation between subjective well-being and depression (rG: approximately 0.8) suggests that polygenic scores derived from GWAS of these different phenotypes may measure the same genetic influence. However, the much lower empirical correlation between polygenic scores (r: approximately 0.2–0.3 in our study) means that conducting the same analysis using different scores could yield different results. Parallel findings across different polygenic scores suggest that it is the overlap among these scores that quantifies the substantive genetic influence. Methods are needed to integrate information from GWAS of different phenotypes with high genetic correlation (
33). In theory, polygenic scores that measure overlap between GWAS results for genetically correlated phenotypes may provide a more precise estimate of genetic influence.
Regarding research on gene-environment interplay, these results highlight two study design issues. First, temporal resolution of environmental exposure measures may be critical. Depression is an episodic condition, including in older adults (
3). Even persistent cases experience temporary remissions. Increases in depressive symptoms following stressful life events may attenuate over time, as is characteristic in bereavement (
34). Our study could account for this attenuation because HRS recorded the date when a respondent died, allowing precise quantification of time since exposure for measures of depressive symptoms. We used statistical models designed for this type of data. This difference may account for why our study could detect gene-environment interplay effects in contrast to some previous analyses (
10,
11). Future studies to detect gene-environment interplay may benefit from focus on stressful life events that can be located in specific temporal relation to the measurement of depressive symptoms (
35).
A second study design issue is the question of whether polygenic scores make good tools for investigating gene-environment interplay in the first place. Polygenic scores have been proposed as a method to investigate gene-environment interplay because, by aggregating information from across the genome, they quantify larger genetic effects compared with individual variants and take on normal distributions (
36). These properties match theoretical and empirical models of genetic influence on quantitative traits (
37). However, it is argued that polygenic scores derived from GWAS naive to environmental variation may quantify primarily genetic influences that do not vary across environments, although alternative GWAS designs are possible (
38). The findings reported above suggest that polygenic scores derived from GWAS naive to environmental variation can predict heterogeneity in response to environmental exposures.
Our study has some limitations. We studied depression in married older adults. The etiology of depression in older adults may differ in some ways from the etiology in younger adults, although the relationship of stressful life events to depression does not seem to vary with age (
39). Etiological features of depression, including subjective well-being, may influence the probability of marriage and of remaining married into later life. Replication of results in younger samples and in unmarried adults is needed. We studied depression following death of a spouse. Depression following death of a spouse may be different from depression related to other stressful life events. However, the distinction is unclear, as is reflected in DSM-5 (
40). There is evidence that so-called bereavement-related depression shares etiology with depression related to other stressful life events (
41). Tests of a well-being polygenic score as a buffer against depressive symptoms following other types of stressful life events are needed. Our analysis was restricted to European-descent HRS participants because of concerns about generalizability of GWAS results across racial/ethnic populations (
18). Studies of genetic buffering against depression following stressful life events in non-European populations are needed.
Research into interplay between genes and environments has a contentious history in psychiatry. With the advent of large-scale GWAS, new, more reliable genetic measures of liability to psychiatric disorders and other mental health characteristics are emerging. At the same time, genetic data are becoming available for large, population-based social surveys that have recorded environmental exposure information about participants. Together, these resources make possible a new generation of molecular genetic research into gene-environment interplay in the etiology of psychiatric disorders.