Craving—the strong desire or urge to engage in motivated action, such as substance use (
1)—is fundamental to addiction (
2) and predicts future substance use and treatment outcomes. More broadly, craving is considered central to understanding motivation for appetitive behavior (
3,
4) and is widely accepted as a construct for behavioral intent in both animal models and humans (e.g.,
5,
6). Self-reported craving has been included as a testing paradigm for constructs (e.g., expectancy/reward prediction error) that are primary to understanding addictive behavior across substance and behavioral addictions (
7). Furthermore, behavior motivated by reward or appetite, mediated by “wanting,” is experienced by individuals with and without substance use disorders (e.g., food craving [
8], non-disordered substance use), with addictive behavior motivated by excessive “wanting” of a substance, or substance craving (
4). There is ample evidence that self-reported craving relates to intake behavior across drugs (
9), food (
10), and other non-disordered craving such as craving for coffee. Therefore, craving represents a target for transdiagnostic modeling that includes comparator samples (i.e., spanning normal and abnormal craving) (
11).
Connectome-based predictive modeling (CPM) is a well-validated machine learning approach for generating predictive models of brain-phenotype associations from whole-brain functional connectivity data, or “connectomes” (
16). In contrast to correlations or regressions, machine learning models, like CPM, protect against overfitting by being defined and validated in independent samples, leading to more accurate effect sizes and increasing the generalizability of findings (
17,
18). CPM is also data driven, and it is therefore used to identify neural “signatures” in functional connectivity data related to a specific phenotype (
16). Importantly, CPM has been used to predict treatment outcomes in substance use disorders (
19,
20), whereas previous approaches had not yet identified reliable brain-based biomarkers of treatment response or abstinence (
21), which is consistent with a neurobiology of successful abstinence involving complex interactions between multiple neural networks (
22).
Methods
Participants
All participants provided written informed consent, approved by the Yale University Human Investigation Committee. Participants included individuals with alcohol use disorder (N=38) or cocaine use disorder (N=28), as determined by the Structured Clinical Interview for DSM-IV and self-report of alcohol or cocaine use, verified by toxicology testing; obesity (body mass index >30; N=19); adolescents with prenatal cocaine exposure (N=41) as well as control subjects (N=148), including adults (N=131) and adolescents without prenatal cocaine exposure (N=17). The participants’ median age was 23 years (range, 14–45 years); 96 were male and 178 were female. Participants were included if they had complete data for self-reported craving and all three fMRI guided-imagery conditions, with an average head motion less than 0.2 mm.
Neuroimaging Data Acquisition
fMRI data were acquired during a personalized guided imagery task in which participants were presented with six imagery scripts: two appetitive (i.e., drug, favorite food), two stress (e.g., romantic break-up), and two neutral-relaxing (e.g., at the beach). Before and after each imagery script, participants rated their craving (their “desire to [use substance (e.g., drink alcohol)] at that moment” or their wanting of the specified food at that moment [“How much do you want (favorite-food) right now?”]) on a scale from 1, “not at all,” to 10, “more than ever” or “extremely high,” thus providing concurrent neural and subjective responses per trial. (For complete task details, see the online supplement.) Participants in studies with food cues were asked to eat ∼2 hours prior to scanning; those with cocaine use disorder were abstinent for at least 2 weeks prior to scanning; all were asked not to consume alcohol for 72 hours prior to scanning; and those with alcohol use disorder were abstinent for 4–8 weeks (for more details, see the online supplement).
Connectome-Based Predictive Modeling (CPM)
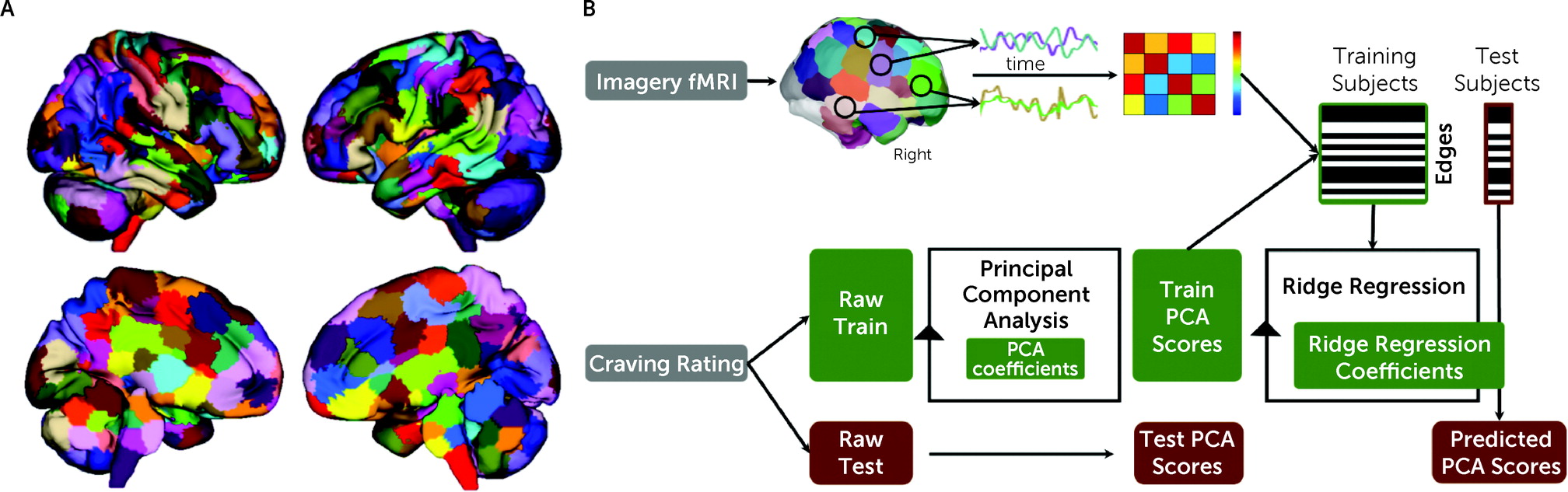
fMRI data were processed for each imagery condition with a validated pipeline and parcellated into 268 nodes using a whole-brain functional atlas (
Figure 1A; see also the
online supplement). Predictive modeling used a multidimensional approach, combining connectomes and craving measures into a single predictive model (
Figure 1B) (
25). A principal component analysis was used to summarize the six craving measures in a data-driven manner for predictive modeling, given that a single behavioral measurement represents a noisy approximation of any behavioral construct, and we did not have an a priori expectation of the best way to combine the measures. Principal component analysis is a common method to reduce the dimensionality of data to increase interpretability and minimize information loss. To maintain separate train and test groups, for each iteration, each analysis was limited to the training data sets and the principal component analysis coefficients were applied to the test data set.
To generate predictive models of craving, all task connectomes were included as independent inputs for a single predictive model using ridge regression (
26). Using multiple connectomes improves predictive modeling and better characterizes brain-behavior associations (
26,
27). However, there is a high degree of similarity between task connectomes, and the edges are not independent. Ridge regression accounts for these dependencies in a principled manner. For feature selection, a statistical significance threshold of <0.05 was used to select edges that are positively and negatively associated with craving in the training data. Potential confounders (head motion, age, group, gender, anxiety, heart rate, smoking status) were controlled for at this step using partial correlation (
25).
Model Validation and Performance
First, to train transdiagnostic models of craving, 10-fold cross-validation was used by dividing the total sample—regardless of diagnostic group—into 10 approximately equal-sized groups; on each fold, the model was trained on nine groups and tested on the excluded 10th group. This was repeated for 100 random divisions. Group membership was not balanced across iterations. Model performance was evaluated with a cross-validated coefficient of determination (q2), and the median q2 for 100 random 10-fold divisions is reported, along with Pearson’s correlation (r), Spearman’s rank correlation (ρ), and mean square error. To generate null distributions for significance testing, we randomly shuffled the correspondence between behavioral variables and connectivity matrices 1,000 times and reran the CPM analysis with the shuffled data. Based on these null distributions, the p values for predictions were calculated as in prior work. Because only a positive association between predicted and actual values can indicate prediction above chance (with negative associations indicating a failure to predict), one-tailed p values are reported.
Second, leave-one-group-out cross-validation was used to test whether prediction results were driven by group differences in prediction performance from including both substance use–related and control groups in a single model. Models were trained on the control group and tested in the substance use–related group and vice versa. To measure prediction performance, Pearson’s correlation was tested between actual and predicted craving.
For additional details on the above methods and quantification of task and anatomical contributions to prediction, see the online supplement.
Group Differences in Connectivity
Group differences in task connectomes (appetitive, stress, neutral-relaxing) between the substance use–related and control groups and how these group differences overlapped with the identified “craving network” were investigated (see the online supplement).
External Validation
Finally, we tested the generalizability of the “craving network” in an independent cohort using fMRI data from six runs of a cue-induced craving task collected before (baseline) and after fasting (
28). This data set consisted of 32 healthy participants who viewed images of individuals talking, laughing, and smiling, highly palatable foods (cake, pizza, chocolate), and attractive flowers (as a control). Participants fasted (except water) for 10 hours before the fasting fMRI scan. Every 2 hours during fasting, self-reported craving was assessed on a scale of 0 to 100, with 100 representing the strongest craving. (For more details, see
https://osf.io/a3yfe/ and
https://openneuro.org/datasets/ds003242/versions/1.0.0.)
fMRI data were processed as described above. One connectome per participant for each of the baseline and fasting scans was created by averaging connectomes for each of the six runs. The five craving measures collected during fasting were averaged to create a single measure of craving. All participants had an average head motion <0.2 mm. Pearson’s correlations between actual and predicted craving and parametric p values were used to measure prediction performance. As above, negative associations indicate a failure to predict, and only positive associations were tested for significance.
Results
Self-Reported Craving
Craving ratings are reported in the Supplementary Results section of the online supplement.
Transdiagnostic Prediction of Craving
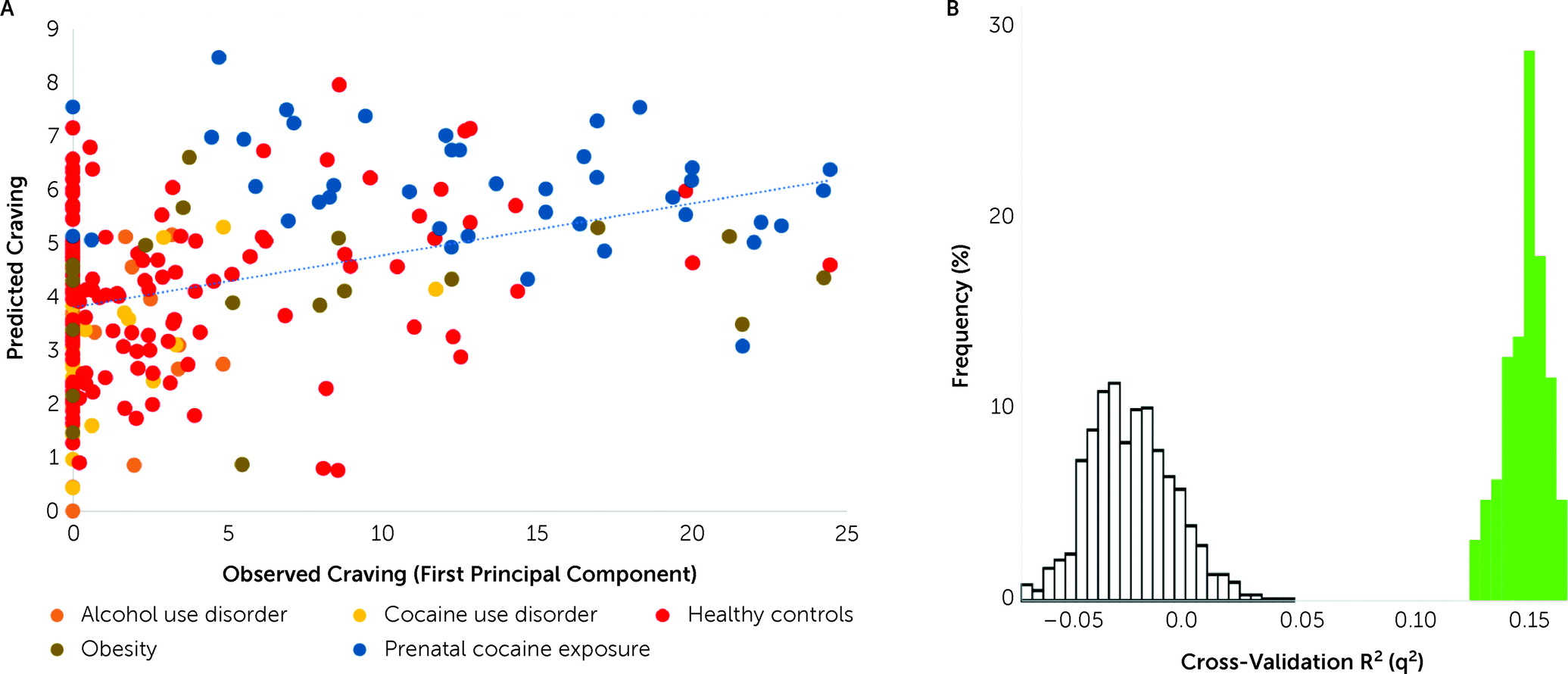
In a transdiagnostic manner, the overall CPM model successfully predicted self-reported craving as defined by the first principal component derived from all craving ratings (r=0.41, ρ=0.37, q
2=0.14, mean square error=34.22, p<0.001, permutation testing 1,000 iterations, one-tailed) (
Figure 2). Follow-up post hoc comparisons adjusting for head motion (r=0.40, ρ=0.36, q
2=0.14, mean square error=34.28), heart rate (r=0.41, ρ=0.37, q
2=0.14, mean square error=34.16), anxiety ratings (r=0.40, ρ=0.37, q
2=0.13, mean square error=34.62), group (r=0.38, ρ=0.36, q
2=0.13, mean square error=34.73), smoking status (r=0.41, ρ=0.38, q
2=0.14, mean square error=34.08), and gender (r=0.41, ρ=0.37, q
2=0.14, mean square error=34.12) demonstrated similar prediction performances. Prediction performance was reduced after adjustment for age but remained significant (r=0.24, ρ=0.24, q
2=0.05, mean square error=37.88). The results were similar when age, gender, and head motion were adjusted for in the same model (r=0.20, ρ=0.21, q
2=0.05, mean square error=38.97). The first principal component explained 92.5% of the variance. Each of the six craving ratings contributed approximately equally to the first principal component (see Figure S2 in the
online supplement). The results were therefore similar when predicting the mean craving, rather than using the first principal component (see Table S2 in the
online supplement). We could not predict imagery-induced changes in craving (post- minus pre-craving; r=0.04, ρ=0.07, q
2=0.00, mean square error=2.5). Finally, models were specific to craving, rather than arousal and alerting to a stimulus, as they did not predict self-reported measures of anxiety (r=0.18, ρ=−0.11, p=0.20) or imagery vividness (r=−0.004, ρ=−0.019, p=0.57).
While the results did not appear to be driven by better prediction in one group (
Figure 2; see also the
online supplement), this was tested by training on the substance use–related group or control group and testing on the other, as the groups differed in craving ratings. CPM predicted craving in the substance use–related group from models trained on the control group (r=0.28, df=146, p=0.001) and vice versa (r=0.19, df=124, p=0.02), showing that prediction was possible regardless of the training data. These two models were also similar, with an edge correlation of r=0.39 (p<0.001) and a node-level correlation of r=0.78 (p<0.001) of model coefficients.
Finally, we tested several alternative models, none of which resulted in superior performance (see Tables S2–S4 in the online supplement), and additionally found no gender differences in prediction performance (see the online supplement).
Anatomical and Task Contribution to Craving Prediction
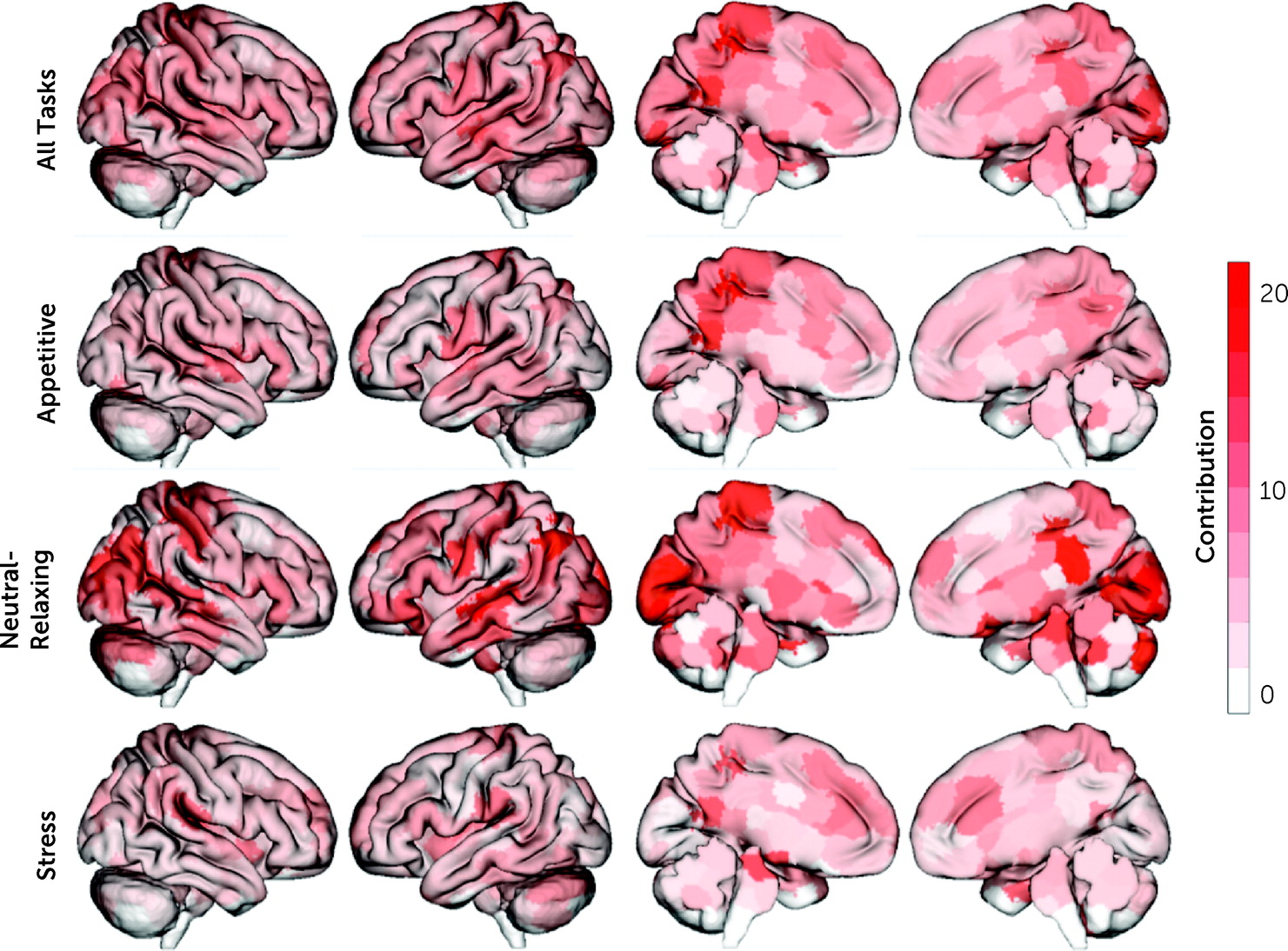
In line with previous CPM results, the identified “craving network” was complex, with contributions from every node and canonical brain network, consisting of 3,069 unique edges (out of 35,778). The top 5% of nodes contributing to craving prediction were located in the posterior cingulate cortex (PCC), hippocampus, visual cortex, and primary sensory areas (
Figure 3). When these spatial maps were uploaded to the Neurosynth Image Decoder (
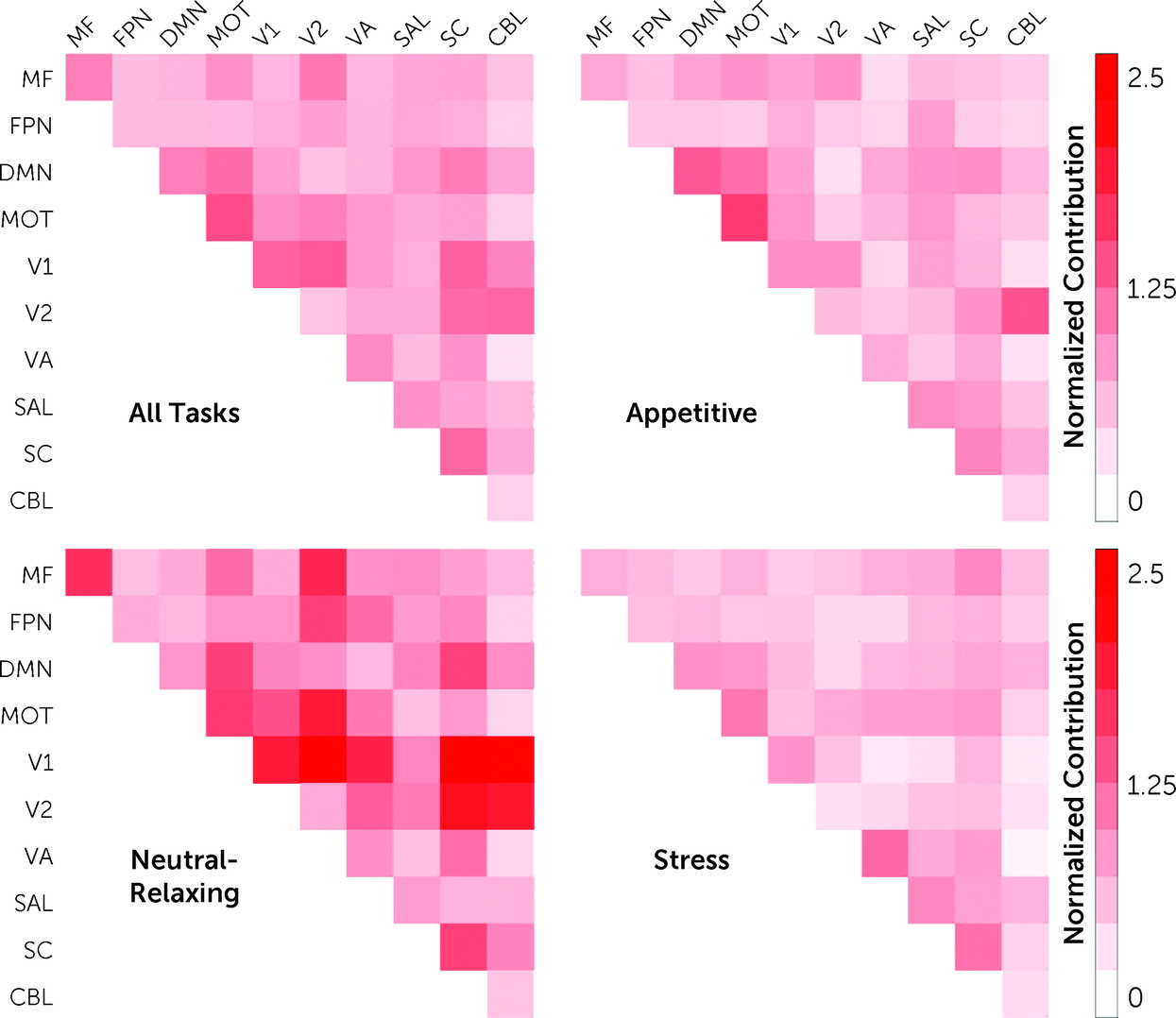
https://neurosynth.org/decode/), “default mode network” was the best match to our “craving network.” Similarly, at the network level, the DMN, motor-sensory, visual, and subcortical networks were the most prominent (
Figure 4; see also Figure S3 in the
online supplement). Finally, virtual lesioning, where canonical networks are iteratively removed from CPM to assess how a single network contributes to prediction performance (see the
online supplement), highlighted that the subcortical, salience, and default mode networks were the most important networks for prediction (see Table S5–S6 in the
online supplement). However, all predictions using a single network were significant (p values<0.01; see Table S6 in the
online supplement).
Next, given that multiple imagery conditions (appetitive, stress, neutral-relaxing) were included in a single predictive model, the contribution of each imagery condition was tested. All task conditions contributed to predicting craving (appetitive, 29.5%; stress, 27.0%, neutral-relaxing, 43.5%), and the distribution of task contributions did not differ significantly from a uniform distribution (χ2=0.05, df=2, p=0.80). For the appetitive condition, nodes with the largest contribution to craving prediction were located in the PCC and primary sensory areas, with the motor-sensory network and DMN as the top contributing networks. Similarly, for the neutral-relaxing condition, nodes with the largest contribution to craving prediction were located in the PCC and primary sensory areas, plus the hippocampus and visual cortex, with the visual networks contributing most to prediction at the network level. In contrast, for the stress condition, nodes with the largest contribution to craving prediction were located in the parahippocampus and brainstem, as well as the PCC, with the subcortical (which includes the brainstem nodes) and motor-sensory networks as the top contributing networks.
Finally, node and network contributions for the appetitive condition were significantly correlated with node and network contributions for both the stress (node: r=0.34, df=245, p<0.001; network: r=0.47, df=53, p<0.001) and neutral-relaxing conditions (node: r=0.35, df=245, p<0.001; network: r=0.31, df=53, p=0.02). However, node and network contributions for the stress and neutral-relaxing conditions were not correlated (node: r=−0.03, df=245, p=0.63; network: r=−0.01, df=53, p=0.95). Overall, model features were similar when adolescents were excluded (r=0.53, df=245, p<0.001).
External Validation
In an independent cohort using a different fMRI task and measures of self-reported craving, the “craving network” successfully predicted self-reported food craving during 10 hours of fasting using both baseline (r=0.47, df=30, p=0.003) and fasting scans (r=0.31, df=30, p=0.04). The difference in prediction performance using baseline and fasting scans was not significant (z=1.01, p=0.31, Steiger’s test) (see Figure S5 in the online supplement).
Discussion
In this study, connectome-based predictive modeling was used to identify a neurofunctional “craving network” in a large transdiagnostic sample of individuals with and without substance use–related conditions. Functional connectivity in the identified “craving network” during an imagery task predicted a continuous measure of self-reported craving across individuals with alcohol or cocaine use disorders, individuals with obesity, adolescents with and without prenatal cocaine exposure, and adult control subjects. Importantly, the neuroanatomical nodes contributing to prediction and group differences were not correlated, suggesting that the “craving network” is neurobiologically distinct from group differences. Finally, for external validation, the “craving network” was used to predict self-reported food craving during fasting in an independent group. These data provide a transdiagnostic perspective to a key phenomenological feature of addictive disorders—craving—and identify a common “craving network” across individuals with and without substance use–related disorders, or across normal and pathological craving, thereby suggesting a neural signature for craving/urge for motivated behaviors.
Consistent with the CPM approach, the “craving network” was found to be complex and distributed throughout the brain. The top nodes contributing to predicting craving included the PCC, hippocampus, visual cortex, and primary sensory areas. The top networks contributing to predicting craving included the DMN, motor-sensory, visual, and subcortical networks. Furthermore, virtual lesion analysis indicated that the DMN, salience, and subcortical networks were the most important networks for predicting craving. These findings are largely consistent with meta-analyses of univariate brain activation studies of substance cue reactivity, which typically implicate the ventral striatum, orbitofrontal cortex, amygdala and hippocampus, middle frontal gyrus, posterior cingulate and parietal cortex, and visual areas (
12), and sometimes the insula (
29), with the amygdala, inferior parietal cortex, and middle frontal gyrus correlated with self-reported craving (
12). The findings are also largely consistent with a study that computed functional connectivity profiles of regions of interest from a cue reactivity meta-analysis to identify subnetworks of cue reactivity–related brain regions and identified visual, executive control, salience, DMN, and an emotion-related network (amygdala, insula, and orbitofrontal cortex) (
14), although that study did not test self-reported craving.
Studies of psychiatric disorders more generally (
30) and of substance use disorders (
31,
32) are increasingly focused on understanding how distributed brain regions organized as large-scale brain networks contribute to dysfunction (
30). The strongest predictor of craving in this study was functional connectivity in the DMN, and the DMN was a match to the “craving network” using the meta-analytic tool Neurosynth (
33). Virtual lesion analysis additionally indicated that the DMN, salience network, and subcortical network were the most important in predicting craving. The DMN is a core functional network (
34), implicated in internally directed thought processes—including self-referential thinking, such as autobiographical, self-monitoring, and social processes (
30)—as opposed to externally directed processing (
35). The salience network is considered to allocate attention to internal or external stimuli (
36). The present finding is therefore consistent with a large-scale brain network model in which a lack of suppression of the DMN, with the salience network allocating attentional resources toward the DMN, results in altered self-referential processing (
30), such as difficulty disengaging from the internal experiences of craving (
23,
32). Mounting evidence suggests that alterations in the DMN are a common feature across psychiatric and neurological disorders (
30,
37), including substance use disorders. We note that the anterior DMN (medial prefrontal cortex) was not found to be predictive of craving. The anterior and posterior midline DMN have distinct roles in self-referential processing, with the anterior DMN implicated in the attribution of personal value and the posterior DMN implicated in internally directed attention (
38). Finally, the subcortical network comprises all subcortical structures, including reward regions (e.g., amygdala, thalamus, striatum), and typically differences in subcortical connectivity between control subjects and individuals with substance use disorders or obesity are interpreted as differences in reward processing (
32,
39). Furthermore, functional connectivity in the salience, motor-sensory, and subcortical networks has been related to reward responsiveness in a model of cocaine abstinence derived using a similar CPM approach (
19). Subcortical structures are also implicated in drug and food craving (
12,
40,
41). Together, the present findings suggest that engagement of the DMN, salience network, and subcortical network in particular represent urge and intent for motivated behavior in response to various appetitive and stress cues that are key contexts that may drive motivated behavior. Increased connectivity in these networks appears to be a common feature of subjective craving that may influence motivated behavior in response to appetitive stimuli across individuals with or without substance use–related conditions.
Transdiagnostic studies such as this can be used to relate heterogeneous phenotypes (wanting/urges/craving) to specific brain networks (posterior DMN, salience network, subcortical network), which can then be related to clinically relevant variations and to genetic, molecular, and cellular factors (
42). Such findings can also be used to evaluate how brain networks are uniquely altered in specific disorders (
20). In this study, participants were grouped as individuals with and without substance use–related conditions, with several subgroups (substance use disorders, obesity, prenatal cocaine exposure) included in the former. Important differences in brain networks related to craving likely exist between these subgroups. However, the purpose of this study was to identify a common “craving network” across groups, or across normative and pathological craving. The diverse set of participants, as opposed to a well-controlled cohort, is needed to show that models are truly transdiagnostic across different subgroups of participants. Furthermore, each subgroup sample size was considered small for neuroimaging-based predictive modeling, where recommended sample sizes are in the range of >100–200 (
43). The study was therefore underpowered for testing subgroups or between subgroups, which remains for future work.
The nodes contributing to prediction and group differences were not correlated, suggesting that the “craving network” is neurobiologically distinct from group differences and may therefore relate to craving/wanting as a more central construct driving human motivated behavior (
4). Notably, the neutral-relaxing condition contributed the most to prediction (43.5%) but showed little between-group difference. This is consistent with transdiagnostic prediction, where the task condition in which groups are the most similar is the most predictive. Transdiagnostic predictive models aim to capture the common neural systems across groups, whereas group differences aim to capture the unique neural systems between groups.
Across both samples, there were numerical, but not statistically significant, differences between craving and connectomes collected at baseline or after cue induction. This may relate to a reported distinction between generalized and cue-induced craving. Generalized or background tonic craving may reflect the incentive or biological value of a drug and may be distinct from phasic, cue-induced craving reflecting drug-cue expectancy (
44,
45). How these components of craving interact is not well understood, but it is likely that they are processed differently. For example, cigarette abstinence has been found to increase generalized craving but not cue-induced craving (
44), and nicotine replacement therapy reduced overall craving but not cue-induced craving (
46). Generalized craving is more similar across individuals with and without substance use–related conditions and might therefore be easier to predict. Likewise, drug cues and abstinence can have separate influences on craving (
45), and drug cue reactivity can be modulated by individual differences in craving in response to abstinence (
47).
Although there is no clear evidence that functional connectivity outperforms fMRI activation or other imaging measures in predicting clinical variables (
48), functional connectivity does offer some advantages for brain-based models. In particular, functional connectivity better reflects the network structure of the brain, which is not represented by activation. Disturbances of network organization are a common thread in virtually all neurological and psychiatric disorders (
49). Because task and rest connectomes are highly correlated, functional connectivity better generalizes to task conditions other than fMRI activation (
50). Finally, methodologically, functional connectivity squares the number of features for machine learning (n nodes leads to n
2 edges), thereby improving power for prediction. Indeed, recent work suggests that connectivity features better relate to phenotypic information than brain structure (
51) and task activation (
52). Nevertheless, these aspects must be tested empirically to assess whether connectivity is a better feature to predict craving than task activation.
This study had some limitations. The craving measure was self-reported and, as with all subjective measures, was therefore sensitive to response bias, introspective abilities or tendencies, and other factors. Some machine-learning studies have found higher prediction accuracy for objective measures than subjective ratings (
53). However, there is no single accepted measure of craving, and nearly all models of craving assume that it can be measured at least in part by self-report (
54,
55). The findings are also generalizable to much of the research on substance craving, which uses self-report (
55). Furthermore, craving was measured in the moment, at multiple time points, and it was highly correlated across time and conditions, thereby reducing potential noise. It would be interesting to test the “craving network” related to objective measures of craving (physiological, behavioral) in future studies, although self-reported craving does relate to intake behavior with food (
10) and drugs (
9). Next, although the results indicated that prediction performance did not differ between genders, and models trained in one gender predicted the other, future studies might test how the “craving network” differs by gender, such as how it is related to clinical outcomes in substance use disorders. Additionally, despite consistencies in how the neuroanatomy of the craving network is situated in previous neuroimaging studies on craving, this literature is mainly based on task activation rather than functional connectivity. These considerations have clinical implications in measuring the “craving network,” with the caution that network activity does not imply the experience of craving. Lastly, while we robustly show that the results were not driven by certain subgroups, there may be effects of other participant characteristics, including clinical and cognitive variables, such as substance use severity and cognitive impairments.
In summary, this study used CPM to identify a transdiagnostic “craving network” in individuals with and without substance use–related conditions. The neuroanatomy of the “craving network” was consistent with previous meta-analyses of cue-reactivity and craving studies, and it critically extends that work to whole-brain functional connectivity using CPM, which is generalizable. This approach indicated that the DMN, the salience network, and the subcortical network were the most important networks in predicting craving. These findings have several potential clinical applications. The DMN has been suggested as a potential biomarker for substance use risk and treatment response and as a treatment target for substance use disorders (
38). The identified “craving network” might be utilized similarly, in particular for individuals who report high levels of craving and for treatments that target craving.