We live in an age where molecular genetics technologies drive rapid advances in our understanding of psychiatric disorders. For example, genome-wide association studies have discovered hundreds of genetic markers associated with mental illnesses in which genes play a major causal role, such as schizophrenia and bipolar disorder. With larger samples, even conditions such as depression—in which life experiences and other risk factors play a larger role in risk than genes—have begun to yield markers (
1–
3) that may ultimately point toward specific genes.
Despite the recent ascendency of molecular methods, we still have much to learn from classical (nonmolecular) methods such as genetic epidemiology, especially when applied to the very large population-based samples that can be studied in settings with wide access to unified health care delivery systems that use electronic medical records. Data from such systems have already breathed new life into the genetic epidemiology of mental illnesses, supporting studies of unprecedented size and scope. Such studies exploit the natural experiments occasioned by families, mate choice, adoption, and twinning to provide more precise estimates of important parameters, including risk to relatives (
4), assortative mating (
5), and heritability (
6).
Heritability is a widely misunderstood concept (
7) that estimates the proportion of individual variation in a trait that can be explained by genes. The rest of the variation, including measurement error, is attributed to noninherited factors. Since it is a proportion, heritability can go up or down in different populations, depending on the degree of individual variation and the magnitude of noninherited factors in those populations. Thus, heritability does not really tell you how “genetic” a disease is but rather how important genes are compared with other risk factors in a given population at a given time. Heritability is traditionally measured in groups of relatives, where genetic relationships can be inferred from the family tree. Twins are a favorite for heritability studies, because identical (monozygotic) twins can be compared with fraternal (dizygotic) twins, providing an excellent control for environmental risk factors shared within families that can confound heritability estimates. More recently, it has become possible to estimate heritability by direct molecular measurement of genetic relationships, typically by use of single-nucleotide polymorphism arrays. These estimates tend to be lower than those obtained from nonmolecular methods, for reasons that remain under debate (
8). Some have suggested that this “missing heritability” is due in part to overestimation of heritability by past twin studies (
9).
In this issue, Kendler et al. (
10) return to traditional nonmolecular methods to estimate the heritability of major depression, relate those estimates to clinical features, and provide new data on the missing heritability problem. Instead of collecting a set of twins or other relatives, the authors instead make creative use of large population-based registries in Sweden. Through these registries, it was possible to examine all treated cases of major depression in Sweden from inpatient, specialist, and primary care settings and select for study not only monozygotic and dizygotic twin pairs but also full and half siblings reared together and apart, totaling more than 1.7 million pairs. This approach provides a larger and more representative sample than would be possible with twin-based ascertainment alone. The siblings reared apart provide an additional control for environmental risk factors, especially half siblings who were separated at an early age.
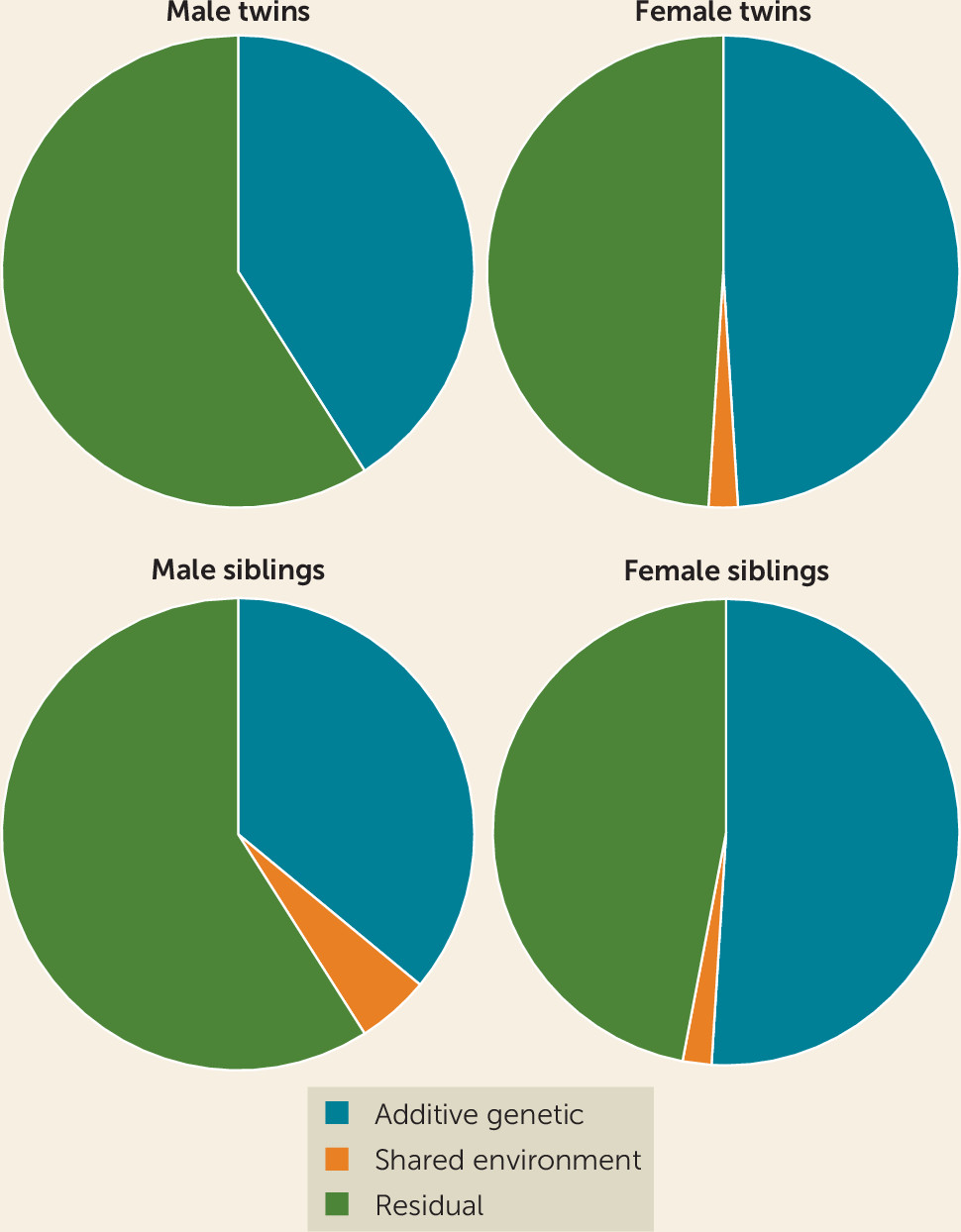
The results showed moderate heritability for major depression. Estimates were around 41%−49% in twins and 36%−51% in the sibling sample, very close to most previous estimates (
11). Heritability was similar in the siblings and half siblings reared apart (36%) and in the twins reared together (41%), suggesting that twin samples do not inflate heritability estimates by much (
Figure 1). Heritability tended to be higher in women, but the majority of genetic risk factors appeared to be shared across both sexes. Major depression was most heritable when characterized by early age at onset, recurrence, comorbid anxiety disorder, and greater clinical severity. Monozygotic co-twins of patients reporting these clinical features were almost three times more likely to suffer from major depression than co-twins of patients reporting none of these features.
This study has many strengths. The sample size is unprecedented for heritability studies of major depression. The findings underscore the continuing value of a good clinical history in assessing patients. The greater heritability observed in patients with early onset, recurrent episodes, comorbid anxiety, and greater clinical severity certainly makes sense and is broadly consistent with the literature (reviewed in reference
11). The finding that most genetic risk factors are shared across both sexes makes sense in light of the highly polygenic architecture of major depression, with numerous alleles of small effect spread over the genome, but it does not shed light on the higher prevalence of major depression often observed in females. More work is clearly needed in this area.
There are also some important limitations to the study design. Only patients receiving treatment were included, so the full range of depressive disorders in the population was not captured. If patients seeking treatment are more severely ill, a treated sample may overestimate heritability, as the authors note. Since the study relies on medical records, clinical features were not systematically assessed, as in studies that evaluate participants for research. There were also no direct measures of adverse life events, a shortcoming of many studies that complicates estimation of the importance of adverse life events in psychopathology. There is also risk of bias when pairs of relatives are raised together by the same parents in the same household. The authors argue that the inclusion of a large number of half siblings reared apart provides a direct test of these shared environmental effects. Time spent in the same household was used to dichotomize the sample into those with and without a shared environment. This is not unreasonable, but it does not take into account the nonlinear relationship between shared environment and depression: adverse events occurring early in life usually have a greater impact than those occurring later. Overall, however, the strengths of the study far outweigh its weaknesses.
Genetic risk for major depression may indeed be reflected in commonly assessed clinical features, but this has been broadly known for decades. The next big challenge is to translate this now quite solid knowledge into a deeper etiological understanding of depressive subtypes and their treatment. As with the common cold, the multiple causes of major depression have frustrated progress in understanding a widespread and common disorder, in this case one that is a major contributor to disability and death worldwide. Defining clinical subtypes that differ in major risk factors would be a great help for clinicians who want to offer patients optimal prognosis and treatment. Such subtypes could also accelerate research into novel treatments, biomarkers, and genetics. The findings of this study should encourage a reexamination of key clinical features as a way to define subtypes of major depression with a larger genetic component. For example, do these same features characterize cases with a greater burden of common risk alleles? Do more heritable forms of major depression show differences in treatment response, illness course, or other important indicators? Can we use genetic information, along with gender, adverse life events, and other risk factors to tailor more effective treatment approaches? These are urgent questions for clinicians and patients. This study shows that they are also the right questions to ask.