Schizophrenia affects about 1% of the general population and is characterized by positive and negative symptoms as well as cognitive deficits (
1). Although our understanding of the genetics related to schizophrenia has advanced (
2), the mechanisms of the disease are still poorly understood and are likely to be diverse. In addition, the current nosology of psychiatric illness is in flux; the concept of a categorical description has been challenged (
3,
4), and even under a categorical nosological scheme, schizophrenia may not represent a single disease entity (
5). Therefore, differentiating subtypes of schizophrenia may be valuable in understanding the mechanisms underlying disease phenotypes. One approach to subtyping schizophrenia is by clinical symptoms or course. However, this may not fully capture the biology that distinguishes patient subtypes. An alternative approach to improving patient subtyping is to identify pathological processes present in subsets of patients.
Abnormalities of protein conformation and solubility have been implicated in many disorders, including cancer, cardiac and pulmonary diseases, muscle diseases, and neurodegenerative disorders (
6). We and others have previously shown that the protein products of genes with rare variants linked to schizophrenia in unique pedigrees are prone to insolubility (
7,
8). Alterations in ubiquitin signaling and ubiquitin proteins have been found in homogenized brain tissue from schizophrenia patients (
9,
10). This is of considerable potential significance because ubiquitin acts as a key modulator of protein insolubility (
11), and ubiquitin reactivity is considered a marker for insoluble protein aggregates in neurodegenerative disorders. Based on these observations, we sought to test the hypothesis that protein insolubility and ubiquitination are present in a subset of brains from patients with schizophrenia and, if these were detected, to determine the potential biological relevance of the insoluble proteins.
Methods
Autopsy Brains, Rat Brains, and Sample Preparation
We conducted a pilot study, a full-scale study, and a replication study in human brain tissue, as well as an experiment in rat brain tissue. For the pilot study, blocks from the prefrontal cortex (Brodmann’s area [BA] 10) of autopsy brains frozen at −80°C were obtained from the Harvard Brain Tissue Resource Center. For the full-scale study, autopsy brains were provided by the University of Pittsburgh Brain Bank. Gray matter was harvested as previously described (
12): tissue slabs containing the superior temporal gyrus, excluding Heschl’s gyrus and planum temporale, were collected in 40-μm sections and frozen at −80°C. A sample of autopsy brains for a replication study was provided by the University of Texas Southwestern Medical Center (
13): tissue slabs containing the prefrontal cortex (BA 46) were collected and frozen at −80°C. These brain regions were chosen for their relevance to schizophrenia (
14–
23). While the brain tissues are not all from the same area because of differences in the availability of tissue from the different brain banks, they represent a pan-cortical sample of brain tissue. For all three brain banks, available covariates were tested for statistically significant differences between patient and control tissues, using unpaired two-tailed t tests for continuous variables and Fisher’s exact test for discrete variables (see Tables S1, S3, and S4 in the
online supplement). Experimenters were blinded to patient or control status during all steps of sample preparation and analysis for all brain cohorts. For the rat brain experiments, male Sprague-Dawley rats were treated with haloperidol (1.5 mg/kg per day), risperidone (6 mg/kg per day), or water only in their drinking water continuously for 4.5 months, as described previously (
24). The rats were then sacrificed by decapitation, and the dorsal striatum/caudate area was dissected and frozen by immersion in −40°C isopentane and stored at −80°C until use. Three rats were used for each condition. Experimenters were blinded to treatment group during all steps of sample preparation and analysis.
Brain Fractionation Protocol
A 10% (weight/volume) homogenate was prepared from human brain tissue in 50 mM Hepes at pH 7.5, 250 mM sucrose, 5 mM MgCl2, 100 mM KCH3COO, 1% Triton X-100, and 1× protease inhibitor cocktail, then centrifuged (20,000 ×g, 20 minutes, 4°C). The pellet was resuspended and centrifuged (130,000 ×g, 50 minutes, 4°C) in 3 mL of high-sucrose buffer (50 mM Hepes at pH 7.5, 1.6 M sucrose, 100 mM KCH3COO, 0.5% Triton X-100, 1 mM phenylmethanesulfonyl fluoride) followed by overnight DNase treatment (100 units/mL) in 3 mL of high-salt buffer (50 mM Hepes at pH 7.5, 1 M NaCl, 20 mM MgCl2, 30 mM CaCl2, 100 Units/mL DNase, 1× protease inhibitor cocktail) at 4°C. The sample was centrifuged (130,000 ×g, 50 minutes, 4°C). The resultant pellet was resuspended and centrifuged (112,000 ×g, 50 minutes, 4°C) in 300 μL of sarkosyl buffer (50 mM Hepes at pH 7.5, 0.5% N-lauryl-sarcosine) to obtain the final insoluble fraction. Each of the above steps was repeated twice. The final pellet was resuspended in 100 μL of 2× Invitrogen NuPage LDS Sample Buffer (56 mM Tris HCl, 70.5 mM Tris base, 1% lithium dodecyl sulfate [LDS], 5% glycerol, 0.255 mM EDTA, 0.11 mM SERVA Blue G250, 0.0875 mM Phenol Red, pH 8.5, 3% 2-mercaptoethanol [BME]).
Western Blot and Quantification Protocol
All insoluble fractions were subjected to SDS-PAGE analysis followed by Coomassie/silver staining and Western blot analysis. The gels were processed on a Criterion TGX 4%−15% precast gel (Bio-Rad Laboratories, catalog no. 567-1084). For Coomassie staining, SimplyBlue SafeStain (Invitrogen, catalog no. LC6060) was used. For silver staining, the SilverQuest Staining Kit (Life Technologies, catalog no. LC6070) was used. Both Coomassie staining and silver staining were used to quantify levels of total homogenate protein and protein insolubility in the final pellet. For Western blot analysis, the gels were transferred to a 0.45 µM PVDF transfer membrane (Immobilon-P, Millipore). A polyclonal anti-ubiquitin antibody produced in rabbit (Dako, code Z0458) was used as the primary antibody for processing the Western blots at a concentration of 1:1000. An anti-rabbit peroxidase-linked secondary antibody (GE Healthcare) was used at a concentration of 1:5000.
Quantification of the protein insolubility was performed by measuring the protein signal of the entire lane for each sample processed on the Coomassie stained polyacrylamide gels and subtracting background signal. The final signal was corrected for the total homogenate protein concentration of each processed sample. The entire area of ubiquitin signal starting from the top of each lane was quantified. The final ubiquitin signal was corrected for total homogenate protein concentration of each processed sample. All gel experiments were repeated with consistent results.
Liquid Chromatography–Mass Spectrometry Data Acquisition
Insoluble pellets were solubilized in 4% SDS 100 mM Tris at pH 7.4 with protease and phosphatase inhibitors (Sigma), digested by trypsin using the FASP method (
25), and analyzed by liquid chromatography–tandem mass spectrometry on an Orbitrap XL Hybrid Ion Trap-Orbitrap Mass Spectrometer (Thermo Scientific) operated in data dependent mode, with a nanoACQUITY UPLC system (Waters) and a picochip column (New Objective), as previously described (
26).
Label-Free Quantification and Protein Identification
All preliminary data processing was performed in Proteome Discoverer, 2.2 beta versions, and finalized in version 2.20.388 (Thermo Fisher Scientific) using an ion current–based label-free quantification method similar to that previously described (
27). Briefly, the total ion chromatogram was subjected to feature alignment using all present ions to compensate for chromatographic shift. Ion traces were stored and matched between runs if they met the post-alignment criterion that retention times within 120 seconds between runs for ions possessing at least three detectable isotopes fell within a 10-ppm mass tolerance. The area under the curve was extracted for the dominant isotope and the trace stored in the output file. Normalization was applied using the total ion current available in each output file and scaled to a total value of 100. All ion abundance measurements were adjusted by this scaling. The normalized protein output was visually inspected to verify algorithm accuracy. The initial analysis contained all pooled technical and digestion controls for further verification of fitness for quantification analysis.
Identification of peptides was performed with SequestHT using a maximum 10-ppm mass tolerance for the parent ion and a 0.6-Da fragment tolerance for tandem mass spectrometry. All data were searched against the cRAP contaminant database (
http://www.thegpm.org/crap/) and the UniProt SwissProt Human canonical database (downloaded on April 10, 2017). Carbamidomethylation of cysteines was considered as a static modification; acetylation of the protein
N-termini and oxidation of methionine were applied as potential variable modifications. Multiple testing correction was performed using false discovery rate calculations, as previously described (
28). A 1% false discovery rate cutoff was applied to both the peptide spectral matches (calculated using Percolator [
29]) and peptide group levels. Quantification ratios for each peptide were determined via pairwise analysis of individual peptides and then averaged for peptide group and protein levels. Significance was then determined by analysis of variance based on peptide background at both the peptide group and protein levels (
30).
Statistical Analysis
Baseline demographic and biochemical characteristics of brain samples from schizophrenia patients were compared with those of control subjects in each of the three brain bank cohorts. Differences between the groups were assessed using Student’s t test for continuous variables and Fisher’s exact test for categorical variables (unpaired and two-tailed). After biochemical analyses of protein insolubility and ubiquitin reactivity, hierarchical clustering analysis was performed on the z-score-normalized concentration of protein insolubility and ubiquitin reactivity in R (hclust function with “average” method; dist function with “euclidean” method; version 3.3.0). After dichotomization into brains with or without increased protein insolubility, the demographic characteristics were compared between patients with increased insolubility and patients and controls without increased insolubility. This was then repeated for patients with increased insolubility compared only to patients without increased insolubility. An unpaired two-tailed t test was used to determine whether the patients with increased insolubility were significantly different from the other patients and controls. The same analysis was applied to ubiquitination data.
Pathway and Cell Type Enrichment Analysis
In an effort to identify dysregulated pathways and cell types contributing to or resulting from protein insolubility, proteins identified by mass spectrometry were analyzed with enrichment tests. Pathways were assessed using gene set enrichment analysis (GSEA; Gene Ontology biological processes gene sets with PANTHER algorithm) and Ingenuity Pathway Analysis (IPA; Qiagen; accessed August 2015). Cell type enrichment was performed using the Expression Weighted Cell Type Enrichment (EWCE) R package (version 0.99.2) and two human cortical single-cell RNA sequencing (RNA-seq) data sets. The first data set, by Darmanis and colleagues (
31), contains expression data for neurons, oligodendrocytes, oligodendrocyte progenitor cells, astrocytes, microglia, and endothelial cells. The second data set, by Lake and colleagues (
32), contains expression data for inhibitory and excitatory neuron subtypes that they identified (N=8 each). EWCE determines the probability of enrichment and standard deviations from the bootstrapped mean for each cell type, using bootstrap significance testing with 20,000 randomly generated gene lists for comparison while controlling for transcript length and GC content. False discovery rates were calculated from the resulting p values using the Benjamini-Hochberg method, and cell types were considered enriched when the false discovery rate was <0.05.
Results
Demographic Information for the Pilot and Full-Scale Studies
For our pilot study, we analyzed samples from five schizophrenia patients and four control subjects from the Harvard Brain Tissue Resource Center matched for age, sex, and postmortem interval (PMI) (see Table S1 in the online supplement). Because this was a pilot study, the demographic data were more limited than for the larger cohorts of the full-scale and replication studies.
For the full-scale study, we analyzed 19 brains from schizophrenia patients (brains S1–S19) and 19 brains from control subjects (brains C1–C19) from the University of Pittsburgh Brain Bank, matched for age, sex, race, PMI, RNA integrity number (RIN), and storage time (see Table S3 in the online supplement). Brain pH was significantly lower in the patient group than in the control group (p=0.006). Patients S4, S6, and S10 died by suicide.
Increased Ratio of Insoluble Protein to Total Protein and Increased Ubiquitination in a Subset of Schizophrenia Brains in the Pilot and Full-Scale Studies
To determine whether protein solubility is disrupted in some patients with schizophrenia, we used two markers of protein abnormality, insolubility and ubiquitination. Protein insolubility is often a consequence of abnormal protein tertiary structure. One of the effects of ubiquitination (addition of ubiquitin to proteins) is to render proteins insoluble (
11). Therefore, to examine protein insolubility and ubiquitination, we performed cold sarkosyl fractionation, an established method (
7,
33–
38) that uses detergent and gradient fractionation to separate soluble and insoluble protein fractions. We quantified the amount of protein insolubility as well as the amount of ubiquitin reactivity in the insoluble fraction. We initially performed this experiment in a small number of brains from the Harvard Brain Tissue Resource Center as a pilot study and determined that a subset of patients with schizophrenia have significant increases in these two markers (see Figure S1 in the
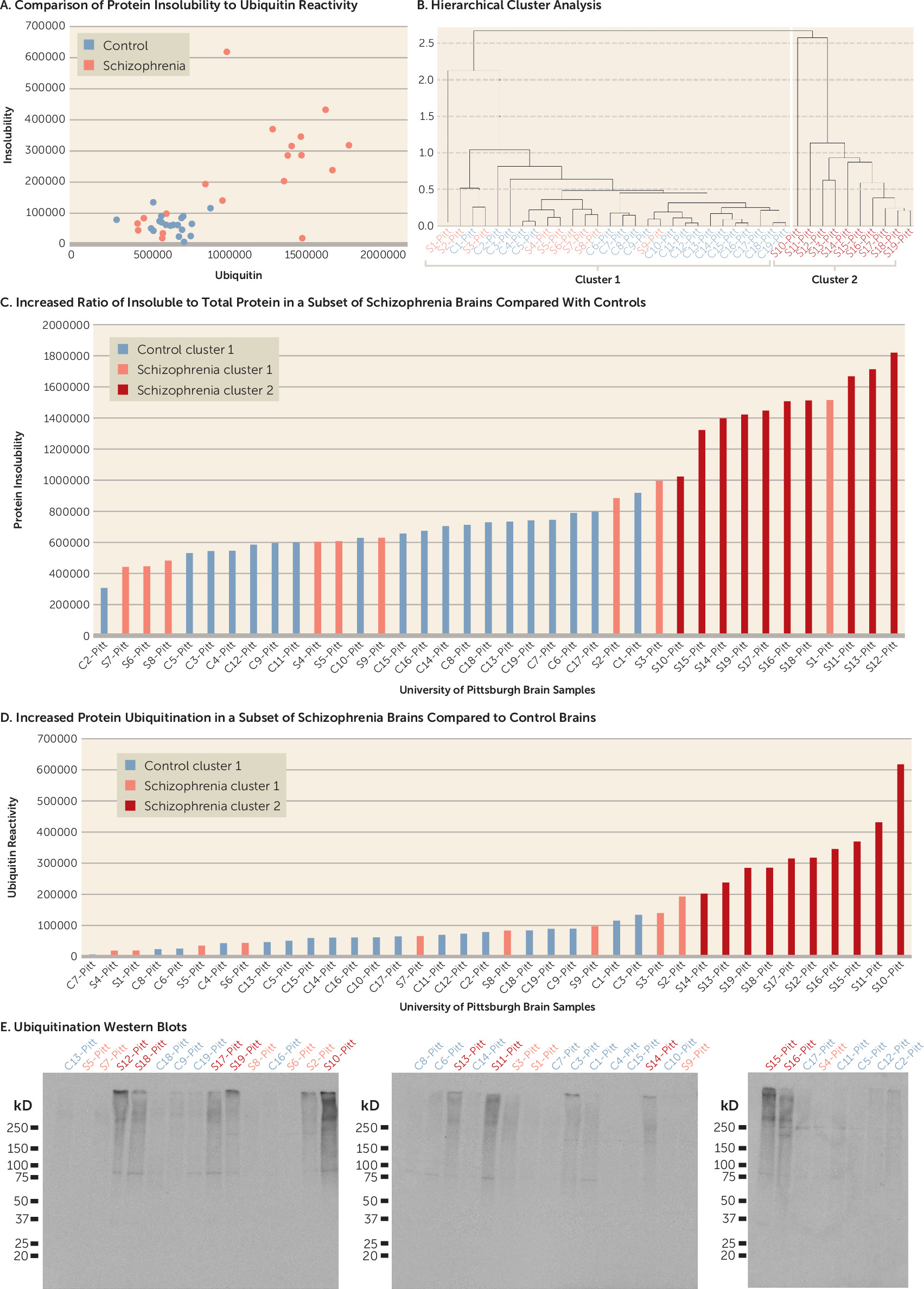
online supplement). Based on these results, we obtained a larger set of brain samples from the University of Pittsburgh Brain Bank. Using this sample, we first demonstrated that joint analysis of protein insolubility and ubiquitination detects a group of brains from schizophrenia patients that appear to separate from all other patients and control subjects based on these two markers (
Figure 1A). Therefore, hierarchical analysis was performed. Unlike flat clustering algorithms, which try to divide a data set into a predefined number of groups, hierarchical clustering yields a tree diagram to show the “natural” grouping structure of the data. Our hierarchical clustering analysis identified two clusters based on protein insolubility and ubiquitination (cluster 1 and cluster 2) (
Figure 1B). We determined that 10 out of 19 schizophrenia brains from the University of Pittsburgh Brain Bank (cluster 2, brains S10–S19) cluster separately from brains of all other patients and control subjects based on protein insolubility and ubiquitination (
Figure 1B). A direct comparison demonstrates that brains in cluster 2 have significantly more protein insolubility and ubiquitination in their insoluble fractions than those in cluster 1 (p<0.001) (
Figure 1C–E). Patient brain S1 was excluded from the positive cluster by hierarchical cluster analysis because it had high levels of protein insolubility but low levels of ubiquitination.
As an additional control experiment, we determined that the total amount of starting protein was not significantly different between the mean of three representative patients with increased protein insolubility and ubiquitination and the mean of three representative controls. We also detected a shift in the amount of protein from the soluble fraction to the insoluble fraction in the patient brains with increased protein insolubility and ubiquitination. When the soluble and insoluble fractions were combined, they added back to approximately the same amount of total starting material, consistent with the presence of an abnormality in protein insolubility in these brains rather than in the total amount of protein (see Figure S2 in the online supplement).
Correlations Between Demographic Characteristics and Increased Protein Insolubility and Ubiquitination in the Pilot and Full-Scale Studies
In our pilot study using the small Harvard Brain Tissue Resource Center sample, differences in age, sex, and PMI between patients with increased markers of protein abnormality and all other patients and control subjects, or between patients with increased markers and patients without increased markers, did not reach statistical significance (see Tables S2A and S2B in the online supplement).
For our full-scale study with brains from the University of Pittsburgh Brain Bank, the differences in age, sex, race, PMI, RIN, or storage time between patient brains with increased insolubility and ubiquitination, compared with all other patient and control brains, did not reach statistical significance (
Table 1). However, the pH in brains with protein abnormalities was slightly but significantly lower (p<0.001). The differences in age, sex, race, PMI, RIN, storage time, or antipsychotic use between patients with increased insolubility and ubiquitination compared with patients without increases in these markers did not reach statistical significance. Brain pH was again slightly lower (p=0.029) and anticonvulsant exposure was higher (p=0.033) in patient brains with protein abnormalities.
Demographic Information for the Replication Study
We obtained a third set of brain samples from the University of Texas Southwestern Medical Center to replicate our findings from the University of Pittsburgh Brain Bank samples. The samples (18 schizophrenia brains, S20-S37, and 18 control brains, C20-C37) were matched for age, sex, race, RIN, pH, PMI, and storage time. As expected, psychotropic medication exposure was higher in the patient samples than in the control samples (see Table S4 in the online supplement). Patients S20, S23, and S32 and control subject C23 died by suicide.
Increased Ratio of Insoluble Protein to Total Protein and Increased Ubiquitination in a Subset of Schizophrenia Brains in the Replication Study
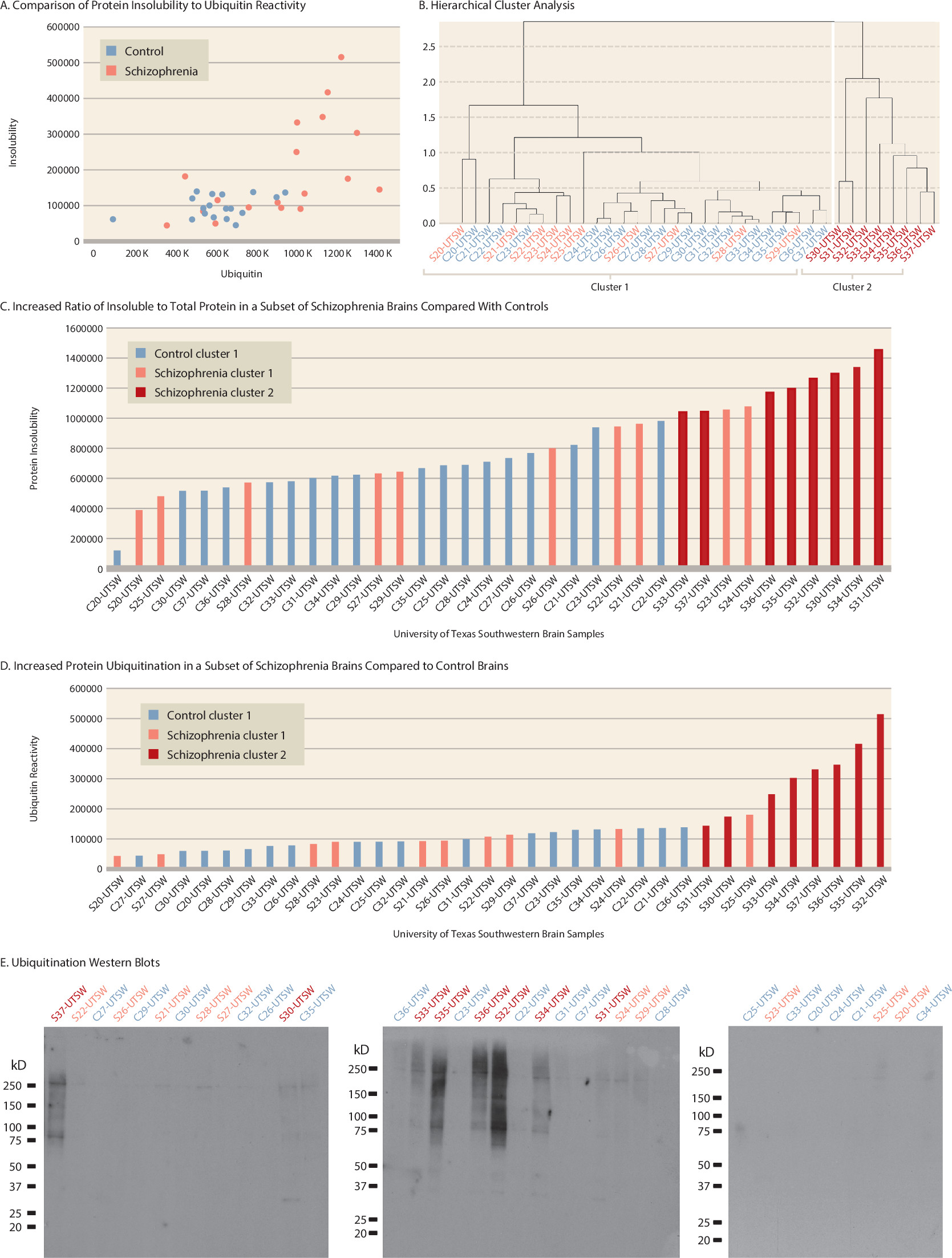
Consistent with our findings in brains from the Harvard Brain Tissue Resource Center and the University of Pittsburgh Brain Bank, eight out of 18 brains from schizophrenia patients (cluster 2, brains S30–S37) clustered separately from the brains of all other patients and control subjects (
Figure 2A and
2B). Again, the brains of cluster 2 tended to have the highest amount of protein insolubility and ubiquitination, and mean levels of protein insolubility and ubiquitination were significantly higher than in brains of cluster 1 (p<0.001 and p=0.002, respectively) (
Figure 2C,
2D,
2E). Hierarchical cluster analysis excluded brains S23 and S24 because they had high levels of protein insolubility but low levels of ubiquitination, and brain S25 because it had a high level of ubiquitination but a low level of protein insolubility.
No Correlation of Demographic Variables With Increased Protein Insolubility and Ubiquitination in the Replication Study
Age, sex, race, PMI, RIN, brain pH, and storage time did not differ between patient brains with increased insolubility and ubiquitination compared either to all other patient and control brains or to brains of patients without increased markers (
Table 2).
Combined Data From the Three Studies
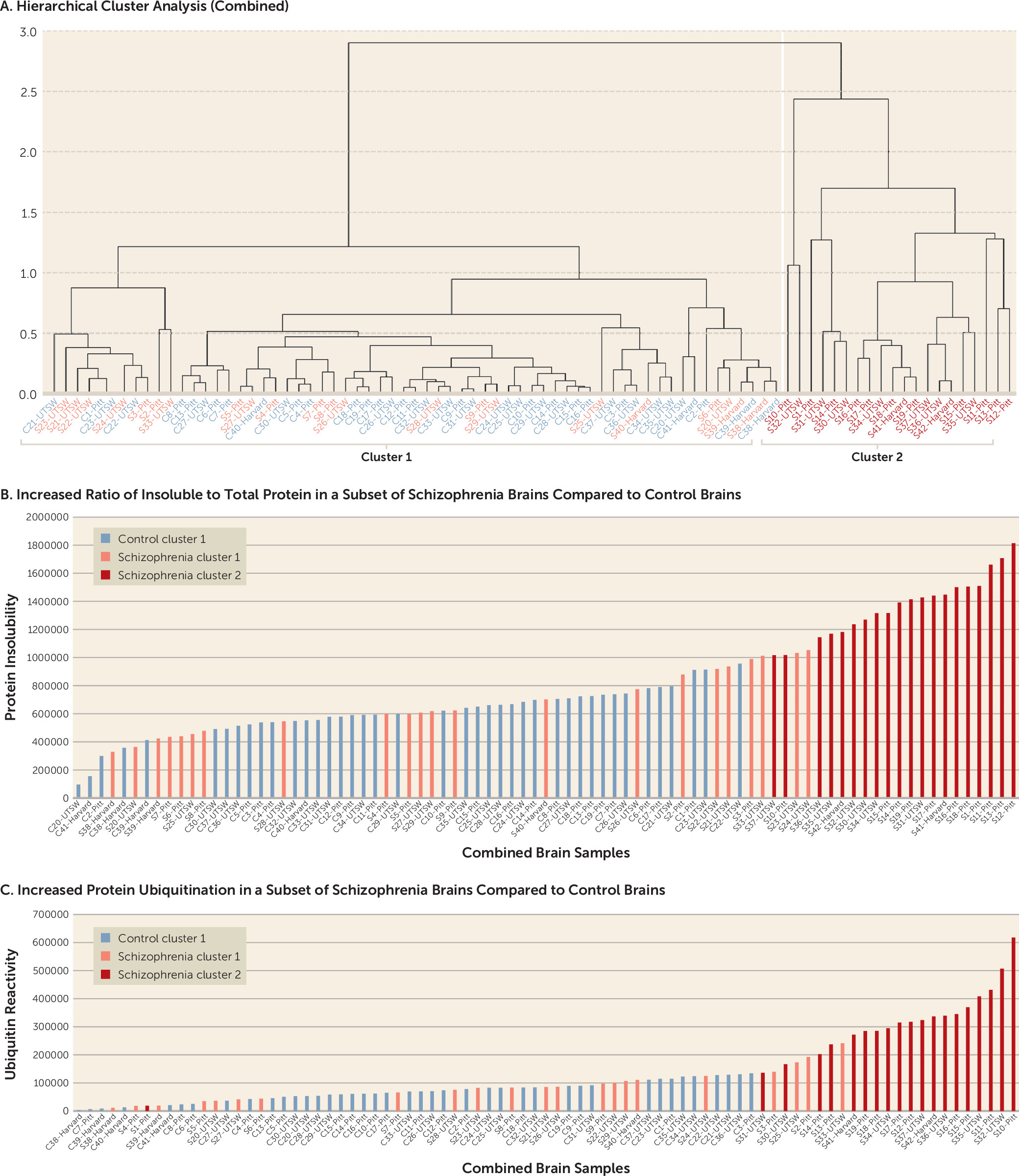
Since insolubility and ubiquitin levels for all three brain sets were normalized to total protein, the data from the pilot, full-scale, and replication studies could be directly combined (
Figure 3). A hierarchical cluster analysis on the combined sample yielded a clustering pattern consistent with the individual brain sets alone (
Figure 3A). In the combined sample, 20 schizophrenia brains had increased protein insolubility and ubiquitination compared with 22 schizophrenia brains and 41 control brains without these abnormalities. The same brains that clustered together in the individual brain sets clustered together in the combined sample, with the exception of brain S1, which switched to the positive cluster, and brain S33, which dropped out of the positive cluster. Brains in cluster 2 had significantly greater protein insolubility and ubiquitination than brains in cluster 1 (
Figure 3B and
3C) (p<0.001 for both).
Correlations Between Demographic Characteristics and Increased Protein Insolubility and Ubiquitination in the Three Studies Combined
While baseline demographic and other characteristics showed a significant difference only in the use of psychiatric medications between patients and control subjects (see Table S5 in the
online supplement), there was a significant difference in brain pH (p=0.002) (
Table 3) between patient brains with increased protein insolubility and ubiquitination and brains of all other patients and control subjects in the combined sample. Furthermore, when comparing patient brains with increased markers to other patient brains, the former were likelier to be female (p=0.019) and to have lower brain pH (p=0.004).
Effect of Antipsychotic Medication on Protein Insolubility and Ubiquitination in Rat Brain
An obvious distinction between patient and control brains is premortem exposure to antipsychotic medication. To determine whether chronic antipsychotic treatment itself can lead to the protein abnormalities detected in a subset of brains from individuals with schizophrenia, we treated rats for 4.5 months with haloperidol (1.5 mg/kg per day), risperidone (6 mg/kg per day), or water alone in their drinking water (dosages were based on previous work showing levels needed to produce vacuous chewing movements in rats). Using the same sarkosyl fractionation protocol applied to human brains, we found no evidence of medication-induced changes in protein insolubility or ubiquitination (see Figure S3 in the online supplement), suggesting that the protein abnormalities detected in a subset of human brains were not a consequence of antipsychotic treatment.
Similar Molecular Signature of Insoluble Proteins in Patients With Increased Protein Insolubility and Ubiquitination
To investigate the potential impact of protein insolubility on cellular function, we performed mass spectrometry for the full-scale study to identify the specific proteins contained in the insoluble fraction from each of the brains in the University of Pittsburgh Brain Bank sample. A total of 182,913 peptide spectral matches were obtained in this analysis, corresponding to 6,958 unique peptides that were condensed to 994 unique proteins. Approximately seven unique peptides were found for each protein identified. (A complete list of the proteins and their quantification values is provided in Table S6 in the
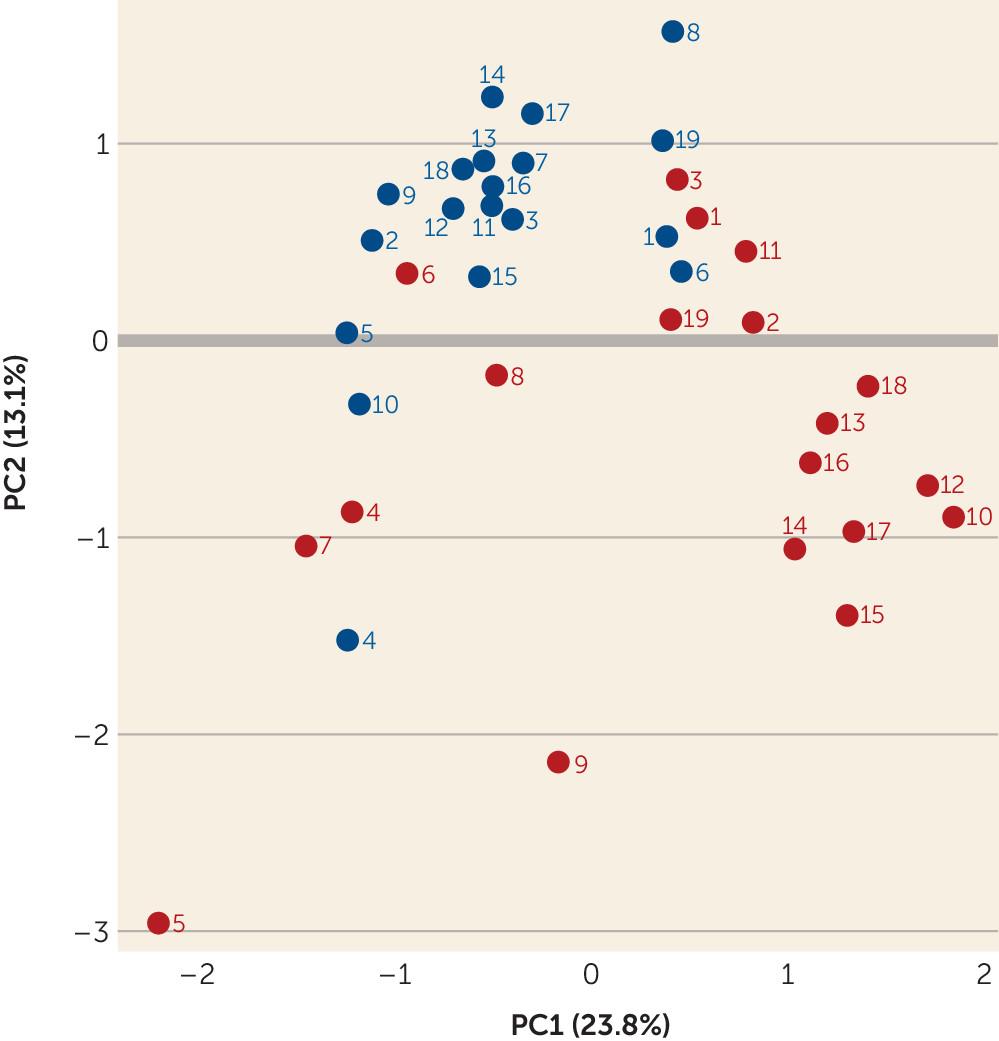
online supplement.) Of these, 602 proteins contained quantification measurements derived from at least two unique peptides for further analysis. A principal component analysis (
Figure 4) demonstrated that the brains with high protein insolubility and ubiquitination separated from the group without these protein abnormalities. Specifically, brains S10, S12, S13, S14, S15, S16, S17, and S18 demonstrated tight clustering, and brains S11 and S19, while less clearly so, appeared to differentiate from the brains of most other patients and control subjects.
Biological Processes and Pathways Identified From the Insoluble Proteins
We next used Gene Ontology (GO) enrichment analysis to analyze all of the proteins that were significantly different between the patients with high insolubility and ubiquitination compared with all other patients and control subjects to identify biological processes associated with the high protein insolubility phenotype. GO analysis identified axon target recognition as the biological process of highest significance (see Figure S4A in the online supplement). We then performed Ingenuity Pathway Analysis (IPA), which provides expert manual curation of pathways and functions to enable the identification of statistical enrichment for a list of proteins in the curated pathways and functions. IPA identified neurological disease as a top pathway for diseases and disorders, cell assembly and organization as a top pathway for molecular and cellular function, and nervous system development and function as a top pathway for physiological system development and function (see Figure S4B in the online supplement).
We also identified proteins enriched in several stages of the ubiquitination process in the insoluble fraction from brains with high insolubility compared with those with low insolubility, consistent with our detection of increased ubiquitination in the insoluble fraction (see Table S7 in the online supplement).
Potential Cell Type Specificity
To determine whether the insoluble proteins are expressed by specific cell types, we utilized single cell RNA-seq expression data from brain cell type groups (from the Darmanis et al. data set) (
31) and neuron subtype groups (from the Lake et al. data set) (
32). We found that the insoluble proteins were enriched in neurons (false discovery rate <0.001) in the Darmanis et al. data set (see Figure S5A and Table S8 in the
online supplement). Analysis with the Lake et al. data set suggested that enrichment occurs in specific neuronal subtypes. Enrichment was significant (false discovery rate <0.05) for one inhibitory subtype (In6) and four excitatory subtypes (Ex1, Ex3, Ex4, and Ex5) (see Figure S5B and Table S8 in the
online supplement). From subtype characterization performed by Lake et al., In6 neurons are parvalbumin GABAergic neurons, Ex1 are cortical projection neurons in layers 2/3, Ex3 are granule neurons in layer 4, Ex4 are subcortical projection neurons in layers 4–6, and Ex5 are subcortical projection neurons in layers 5–6. We recognize that studying proteins through their RNA expression has its limitations. Further analysis should be performed when a brain cell type expression database is available for protein expression.
Discussion
In this study, we identified a subset of brains from schizophrenia patients with increased protein insolubility and increased protein ubiquitination in cortical tissue. When the insoluble fractions were analyzed by mass spectrometry, we found proteins that were present in all brains but found in greater abundances in the brains with high insolubility and ubiquitination. We also found unique proteins that were detected only in brains with high insolubility and ubiquitination. The insoluble proteins are disproportionally reflective of pathways related to nervous system development and axon target recognition, including axonogenesis, neurogenesis, and neuron projection.
Given the novelty of our findings, we used a tiered strategy of examining a small number of brains from the Harvard Brain Tissue Resource Center to explore our hypothesis, then retesting the hypothesis in a larger sample of brains from the University of Pittsburgh Brain Bank, and replicating the findings in a third sample of brains from University of Texas Southwestern Medical Center. Each sample yielded the same result, as did analysis of the combined samples, increasing our confidence that increased protein insolubility and ubiquitination are not artifacts related to the ascertainment, processing, or storage of postmortem brains in a particular brain bank. Furthermore, the available tissue from each brain bank was from a different cortical region, yet results were similar, consistent with the idea that aspects of schizophrenia pathophysiology have pan-cortical involvement (
39–
43). In addition, we used three different but related markers to explore protein abnormalities: insolubility, ubiquitin, and peptide abundance. As a result, we detected increased levels of these markers in a cluster of the same brains. We posit that this cluster identifies a subset of patients with a common pathophysiology.
Analysis of autopsy brains presents a number of challenges. Brain pH may reflect postmortem changes and storage conditions, and it could influence protein properties. We detected lower pH in the brains with protein abnormalities in the University of Pittsburgh Brain Bank sample and the combined sample from three brain banks, but not in the University of Texas Southwestern Medical Center sample analyzed alone. However, some of the brains with increased protein insolubility and ubiquitination were within the same pH range as brains from schizophrenia patients and control subjects without protein abnormalities. In addition, RIN values, which may be a better proxy for brain quality (
13), did not differ significantly in any of our group comparisons. Interestingly, a difference of 0.2 to 0.4 pH units is frequently detected between schizophrenia and control brains (
44–
47). In addition, a magnetic resonance spectroscopy study showed that schizophrenia patients have significantly lower intracellular brain pH compared to control subjects (
48). While the reason for this difference is not fully established, it may be related to premortem alterations in metabolic pathways and lactic acid (
46,
49–
53). In addition, rodent models indicate that oxidative stress can lower brain pH (
54–
56). Importantly, both processes are stress cascades implicated in schizophrenia; it is therefore possible that the processes leading to lower pH in schizophrenia brains is a primary intrinsic pathogenic process related to protein insolubility.
In general, alterations of protein conformation and solubility have been implicated as part of the primary pathogenic process (rather than a consequence of medication) of many disorders, including cancer, cardiac and pulmonary diseases, muscle diseases, and neurodegenerative disorders (
6). However, antipsychotics and other medications influence various biological processes in the brain, potentially including protein processing. To address the concern that antipsychotics may influence protein solubility and ubiquitination, we chronically treated rats with either haloperidol or risperidone, representing two different classes of antipsychotics. Neither treatment led to protein insolubility or ubiquitination, suggesting that exposure to antipsychotics did not generate the protein abnormalities that we detected in human brains. However, this experiment was done without the extended PMI found in the human cases. In our data sets, there was no significant difference in exposure to antipsychotics, antidepressants, or benzodiazepines between brains with and without increased protein insolubility or ubiquitination. Exposure to anticonvulsants was significantly associated with increased protein insolubility and ubiquitination in the University of Pittsburgh Brain Bank samples, but not in the University of Texas Southwestern samples. However, there was no overall significant difference in anticonvulsant exposure when samples from all brain banks were combined. Anticonvulsants, and in particular valproic acid, may have neuroprotective properties in neurodegenerative disorders characterized by protein insolubility (
57) and therefore likely do not promote protein misfolding and insolubility. Together, these data suggest that protein insolubility is intrinsic to the mechanism of a subset of mental illness and is not related to medication effects.
Since the brains in all three of our samples were obtained from coroner’s offices, there was limited availability of detailed social, medical, and psychiatric history to link abnormal protein solubility to a specific clinical phenotype, familial factor, or environmental exposure. However, there was a significant sex difference, with protein insolubility and ubiquitination more frequently found in brains from female patients in the combined sample. It is unknown whether this finding can be replicated in other brain collections and what the potential biological implications are. Furthermore, limitations in sample size precluded an analysis of the correlation of protein abnormalities with substance use or suicide; however, these phenomena were not prevalent in our samples and were nearly evenly distributed in brains with and without protein abnormalities. We suspect that cigarette smoking, whether through nicotine or other factors, is unlikely to contribute to individual variation in protein insolubility, as smoking was common to both patients with and without protein abnormalities, but we cannot exclude the relevance of variation in tobacco exposure with regard to protein abnormalities. Future analyses of brain samples with more clinical history would help clarify these issues.
Our findings are consistent with proposed pathogenic mechanisms of schizophrenia. While schizophrenia is not associated with cortical neuron loss (
58), sequestration of proteins into insoluble forms could lead to disruption of critical pathways, resulting in neuronal dysfunction. Interestingly, while protein misfolding is often associated with neuronal death in neurodegenerative disorders, our pathway analysis implicated a neurodevelopmental role for protein insolubility in schizophrenia, consistent with the neurodevelopmental hypothesis of this disease (
59–
61). Enrichment of insoluble proteins in axon target recognition and neuronal projection pathways may be consistent with the related hypothesis that schizophrenia is a consequence of aberrant development of neuronal projections and connectivity (
39,
62,
63).
A role for protein insolubility in schizophrenia has previously been suggested by studies of rare genetic variants associated with this disease (
7,
8). DISC1, a protein originally associated with schizophrenia and affective disorders through a chromosomal translocation in a single large family, was identified in the insoluble fraction in a subset of brains from individuals with depression, bipolar affective disorder, and schizophrenia (
64). Also, in a rat model, DISC1-containing protein aggregates disrupted dopamine homeostasis (
65). A mutation in the neuronal transcription factor NPAS3, linked to schizophrenia in a small family (
66), leads to the aggregation of NPAS3 and interferes with its transcriptional functioning (
8). Other genetic factors associated with schizophrenia have been linked to similar protein abnormalities, including dysbindin-1, CRMP1, and TRIOBP-1 (
67–
70). Furthermore, environmental insults related to oxidative stress or altered immune signaling are linked with neuropsychiatric disorders and have also been associated with protein misfolding, such as endoplasmic reticulum stress and heat shock factors (
71,
72).
Overall, our results provide strong support for the hypothesis that protein insolubility occurs in a subset of schizophrenia patients. Based on the function of the detected proteins, we suggest that the insolubility is related to disease pathogenesis. Future experiments could refine the regional and cell-type specificity of the phenomenon and elucidate the possible relationship between protein insolubility and other psychiatric disorders (e.g., bipolar disorder and major depressive disorder). Further exploration of the molecular mechanism underlying the protein insolubility in subsets of patients with schizophrenia or other disorders could lead to a better understanding of the pathways, circuitry, and symptoms seen in patients with mental illness, and could lead to improved nosology and novel therapeutic targets.