“Every human being comes to earth with sealed orders.”
—Søren Kierkegaard
Suicide, terrible and terrifying, is surprisingly common. However, viewed temporally, death by suicide is a rare, and indeed once-in-a lifetime, event. Adding to the mystery of suicide, no other animal consciously devises its own death. Suicide is feared because of its unpredictability and stigmatized in part for the same reason, but also because of the damage it leaves behind. For those grieving suicide victims, graveyards, contrary to de Gaulle’s ironic aphorism, are full of irreplaceable people. Prediction and an understanding of the causes of suicide can save lives and inform us about disease conditions that sometimes lead to suicide but would also teach us to understand, and in other ways to improve, the human condition.
As may seem paradoxical, the likelihood of suicide is partly heritable. Our fates are not sealed, in Kierkegaard’s terms, but are influenced by inborn factors. In this issue of the
Journal, Docherty et al. (
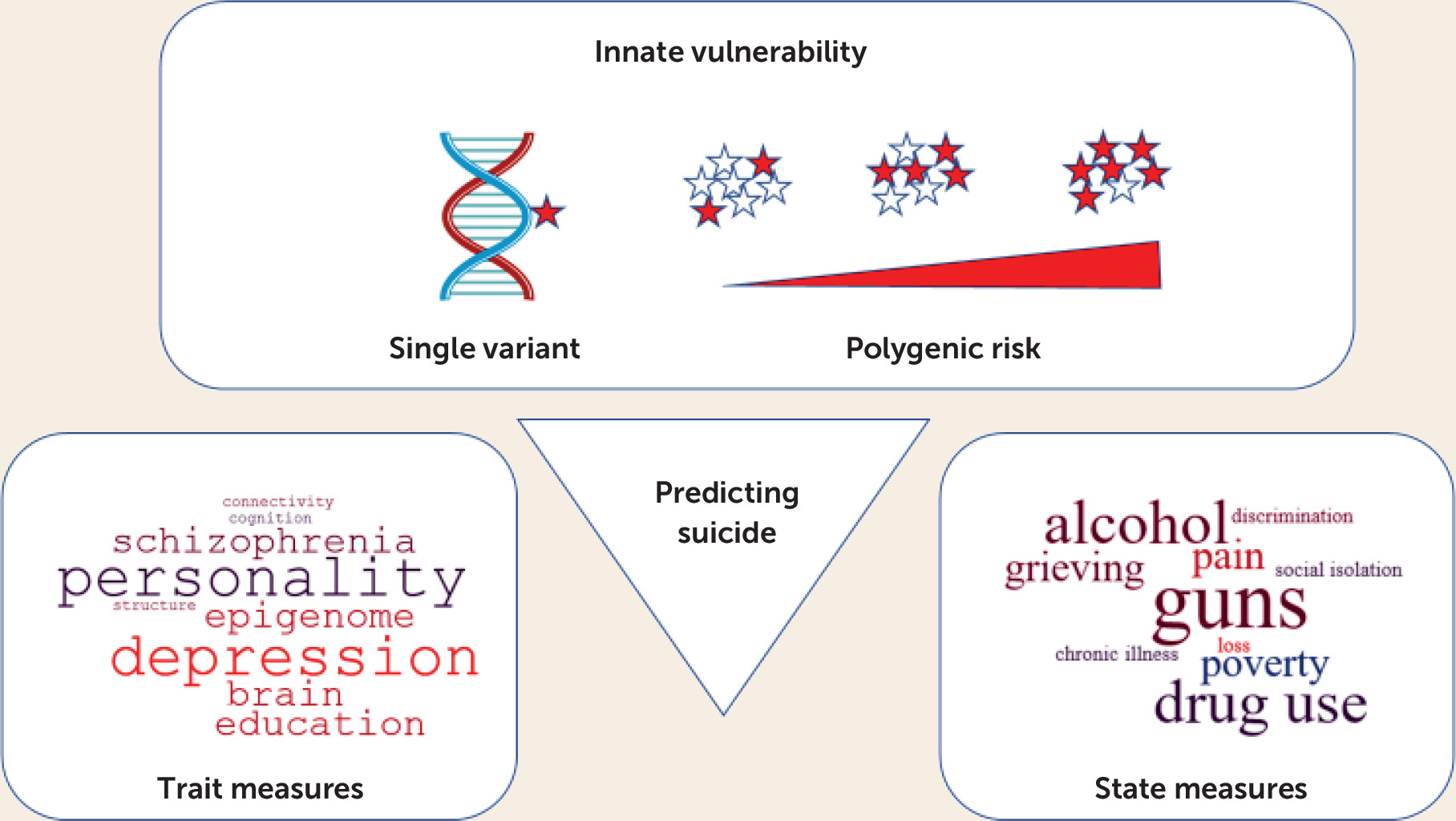
1) took a new approach to the prediction of suicide and its clinical antecedents, adding to the elements that might be combined, perhaps via machine learning, to predict suicide (
Figure 1). They performed a genome-wide association study (GWAS) on suicide, rather than suicidality, which might also uncover different genes. Their GWAS, of more than 3,400 suicide victims from the state of Utah and several times that number of ethnically matched comparison subjects from outside the state, implicates two genes that were genome-wide significant and many more genes that were significant by gene-wise analysis. Gene-wise GWASs (see Liu et al. [
2] for example), take advantage of multiple single-nucleotide polymorphisms (SNPs) genotyped at most genes on arrays used for GWAS and integrate this information to reduce multiple testing, increase statistical power, and facilitate gene-network analyses. The identification of genes that are significantly associated with suicide, and the implication of a much larger number of other genes in the genomic statistical threshold, enabled Docherty et al. to derive a polygenic risk score accounting for a substantial portion of the risk of suicide. Based on estimates that suicidality is 17%−55% heritable, they may have found an even larger portion of risk attributable to genes. Shedding light on why genes contributing to suicide exist, the authors found genetic cross-connections to other psychiatric diseases and behaviors, including behavioral disinhibition, depression, psychosis, autism, and alcohol use disorder. As a species, humans are inherently diverse in behavior, making us stronger as a community. But we also pay a price in individual vulnerability to psychiatric diseases. Identification of genetic risk factors can target interventions to enhance lives—for example, by preventing suicide—without diminishing our essential diversity or reducing the origins of suicide to a simplistic, deterministic formula.
With the Docherty et al. study, and other genomic studies, we are now at the “beginning of the beginning” in understanding genetic vulnerability to suicide. A main lesson of GWASs is that larger samples, and samples more deftly measured for phenotypes strongly influenced by genes, yield more genes and may yield findings that are more durable. Docherty et al. were mainly unable to replicate findings from smaller suicide and suicidality genomic studies (as they reviewed), as well as from a larger study based on the UK Biobank and Vanderbilt University (BioVU) electronic health records data sets (
3), from a larger study based on the UK Biobank data set only (
4), and from a larger study conducted in Denmark (
5). This lack of replication is unsurprising because at this point, only a small percentage of the variance is accounted for by most GWASs of suicidality—the polygenic heritability was 15% in the Utah study by Docherty et al. (
1) but 4% in the UK Biobank and BioVU studies by Ruderfer et al. (
3), 7.5% in the UK Biobank study by Strawbridge et al. (
4), and 4.6% for the Danish sample in the study by Erlangsen et al. (
5). In these studies, there are no, or few, marginally significant loci, and the phenotypes vary from deaths by suicide, as reported by Docherty et al., to measures of suicidality on questionnaires of well-being.
No finding convergent with an earlier generation of studies on candidate genes implicated by the neuroscience of depression and impulsive behavior has yet emerged from GWASs. Notably, for other psychiatric phenotypes, convergences between neuroscience and GWASs are now commonplace, and, for example, the D
2 dopamine receptor, a major drug target in psychosis, is implicated in schizophrenia, and already at a p value of 10
−12, and with associations as strong as 10
−40 for depression and anxiety, alcohol and drug use, and sense of well-being. However,
DRD2 has not been linked to suicide, as would be expected because of linkages to suicide-related phenotypes. Concerning that early generation of studies, but leading directly to genetic testing being performed today in psychiatry, a hypothesis that never dies is that depletions of brain serotonin and, as reflected in low levels of the metabolite 5-HIAA in CSF (
6), serotonin receptor binding potential (
7), as well as serotonin itself (
8), predict suicide. Genes such as the serotonin receptor
HTR1A, implicated by Mann and colleagues, and
TPH1 and
TPH2, encoding tryptophan hydroxylase, the rate-limiting enzyme for serotonin synthesis, are again missing in action for suicide GWASs and psychiatric GWASs overall. Naturally, these genes were objects of candidate gene studies (
9) and searches for functional variants (
10), but what remains lacking is any signal of genetic association to behavior that would survive genome-wide correction. The only phenotype yet linked to
TPH1 (and barely significant) is high-density lipoprotein cholesterol, and there is no significant GWAS linkage of any kind to
TPH2. Also, it would seem plausible that
SLC6A4, encoding the serotonin transporter, would predict depression and suicide because the serotonin transporter is a major target engaged in the treatment of depression, as well as because expression of
SLC6A4 is altered by a common polymorphism. However, genetics is usually more complicated than it superficially appears. The
SLC6A4 polymorphism is a short tandem repeat variant, with at least three common functional alleles (
11) that of course would not be fully interrogated by individual biallelic SNPs on GWAS arrays. The top GWAS hit for
SLC6A4 is body mass, and the serotonin transporter has never been implicated by GWASs to any behavior. Whether GWASs of suicide will eventually be linked to genes such as
DRD2 and
SLC6A4 is an intriguing question, and if not, why not?
Do any of the candidate genes, or polygenic combinations, offer a tool for clinical prediction at present? Here, it is important to distinguish between genes altering drug metabolism, where we are further along, versus genes that predict disease risk, and where we have far to go. The National Institute of Mental Health has issued a fact sheet on the utility of genetic testing, including consumer testing, in mental health (
12). As of now, “the short answer is no” regarding whether genetic testing will help to predict risk. Whether a few genes, or many, are used to predict, more foundations are needed for genetic tests for suicidality, or to predict diseases such as depression that trigger suicide (
13).
However, taking a longer view, these beginnings are encouraging. GWASs such as the one by Docherty et al. have already taken us beyond the genes and pathways that were “under the lamppost” to new targets for mechanistic studies, and these genes in combination appear to predict risk attributable to multiple variants of small effect. Presently, physicians collect information on clinical risk factors and very directly and sensitively ask about feelings and concreteness of plans, integrating that information into a risk profile and treatment. The ability of geneticists to capture increasing portions of the heritability of suicide may enable psychiatrists to better help people at risk, combining genetic predictors of innate (trait) vulnerability with such state and trait measures.