Schizophrenia affects approximately 1% of the general population if considered as a spectrum of genetically related clinical diagnoses
(1). Despite numerous biological studies, no underlying inherited mechanism has been found. As polymorphic DNA markers and the laboratory techniques for high throughput linkage analyses have become available over the last 20 years, application of this strategy to finding genes for schizophrenia has been rigorously pursued by several research laboratories. However, numerous reports suggestive of linkage to specific chromosomal regions have contributed to widespread uncertainty concerning the significance of these diverse findings
(2–
13). Most recently, there have been reports of significant linkages on chromosomes 13q and 1q
(5–
7,
13–15). Despite all these efforts, at present there have been no convincing reports of any mutation in a schizophrenia susceptibility gene that demonstrate a high degree of statistical significance and/or an alteration in function.
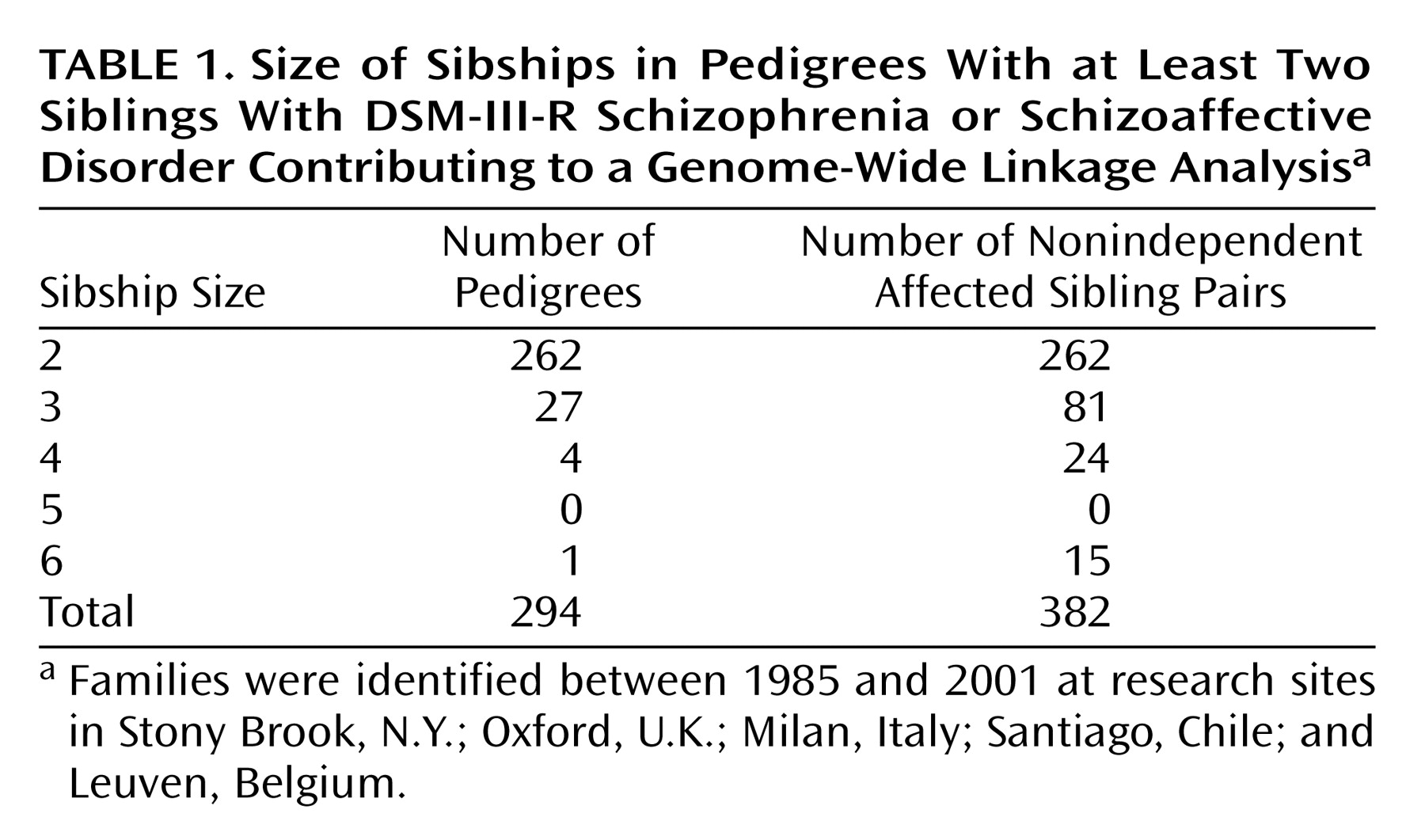
We previously reported a genome-wide scan performed with 70 families having at least two affected siblings, at least one of whom had a diagnosis of schizophrenia
(16). No genome-wide significance was observed, but lod scores above 2.0 or a p value of <0.01 were seen for chromosomes 1q22-q23, 2p14-p13, 4pter-cen, 5p15, 10q22-q24, 11pter-p11, 12p13-q24, 13q12-q13, 16q22.1, and 22q11. The present study was an expansion of the previous genome screen with the number of families increased to 309. We previously published negative results for this cohort on chromosomes 8, 13, and X, in response to earlier reports of linkage on these chromosomes
(17,
18). The present manuscript summarizes the entire genome-wide scan and reports one significant and one suggestive region of genome-wide significance, according to the criteria for linkage of Lander and Kruglyak
(19), but fails to confirm a number of previous reports of linkage. The present findings suggest that a critical reevaluation of the linkage approach is warranted.
Results
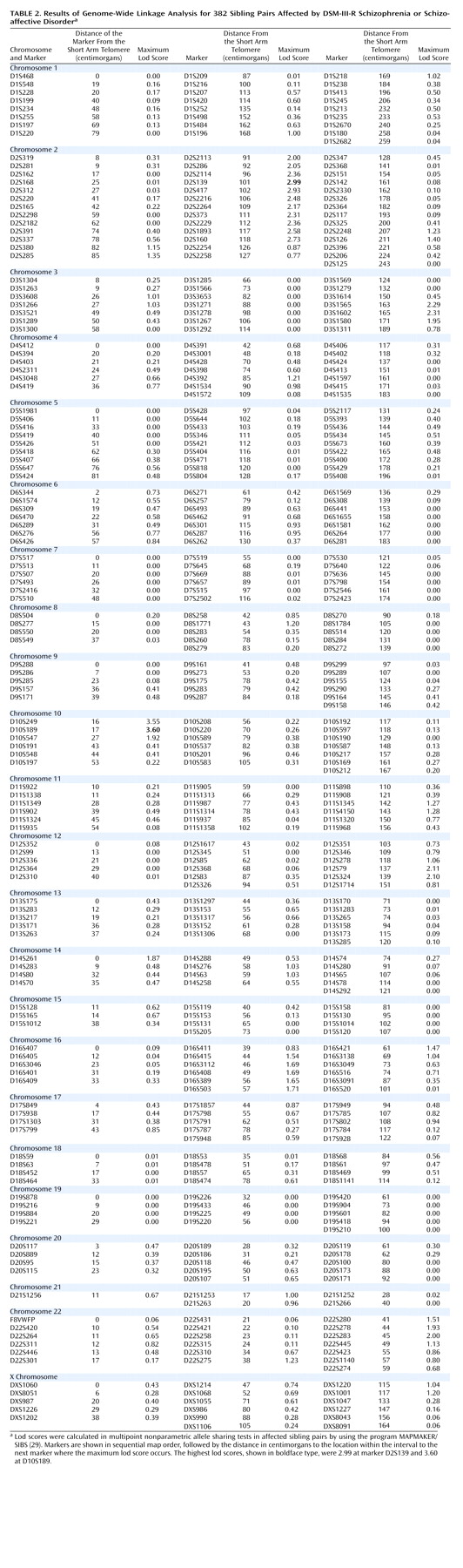
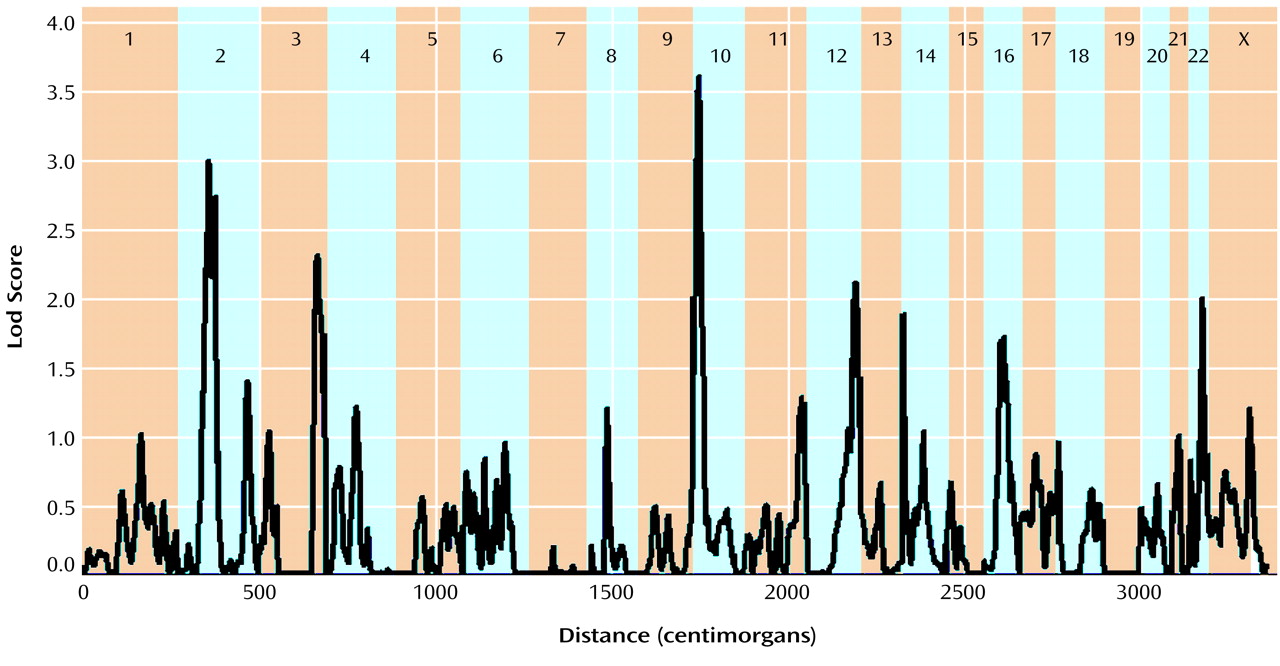
The multipoint maximum lod score between each marker interval is indicated for all markers included in the genome scan (
Table 2). There was one region with significant and two with suggestive linkage in this genome scan (lod score greater than 2.2
[19]). The significant peak lod score occurred in 10p14 at marker D10S189 with a Z
max of 3.60. The second best linkage result occurred in the centromeric region of chromosome 2 with a peak lod score of 2.99 between markers D2S139 and D2S417. A second peak on chromosome 2 occurred approximately 17 cM distal in 2q12 between markers D2S160 and D2S2254, with a peak lod score of 2.73. Finally, the third best linkage result occurred in 3q27 between markers D3S1602 and D3S1580 with a peak lod score of 2.31. Other peak lod scores ≥2.0 were obtained on chromosome 12q22-q24 at marker D12S324 (Z
max=2.10) and on chromosome 22q12-q13 at D22S283 (Z
max=2.00). The multipoint maximum lod score curves for all chromosomes are shown in
Figure 1. All of the corresponding lod scores and markers used are shown in
Table 2.
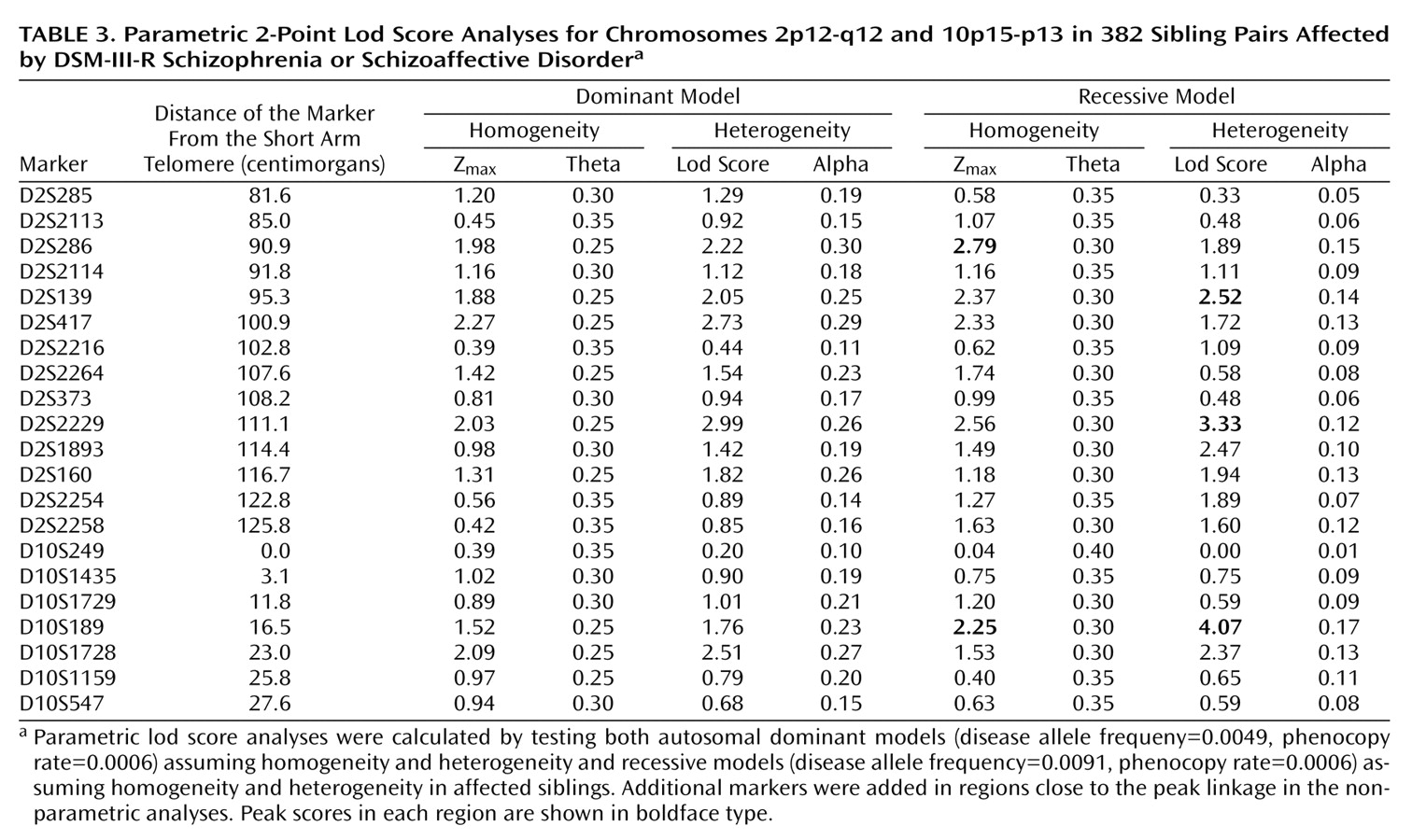
Parametric lod score analyses were calculated for markers in the chromosome 2 and 10 linkage regions, testing both autosomal dominant and recessive models by using an affecteds-only analysis. The results are shown in
Table 3. In the chromosome 10p region, the peak parametric lod scores were obtained by using a recessive model (disease allele frequency=0.0091, phenocopy rate=0.0006). The peak lod score under homogeneity occurred at D10S189, with a Z
max of 2.25 (theta=0.30). Under heterogeneity, the lod score increased to 4.07 at D10S189 (theta=0.00, alpha=0.17). In the centromeric region of chromosome 2, the highest lod score assuming homogeneity occurred at D2S286, with a Z
max of 2.79 (theta=0.30), also by using a recessive model. When the data were analyzed under heterogeneity, the highest lod score occurred at D2S2229, with a lod score of 3.33 (theta=0.00, alpha=0.12). In addition, under heterogeneity, marker D2S139 located 16 cM proximal to D2S2229 in 2q12 had a peak lod score of 2.52 (theta=0.00, alpha=0.14).
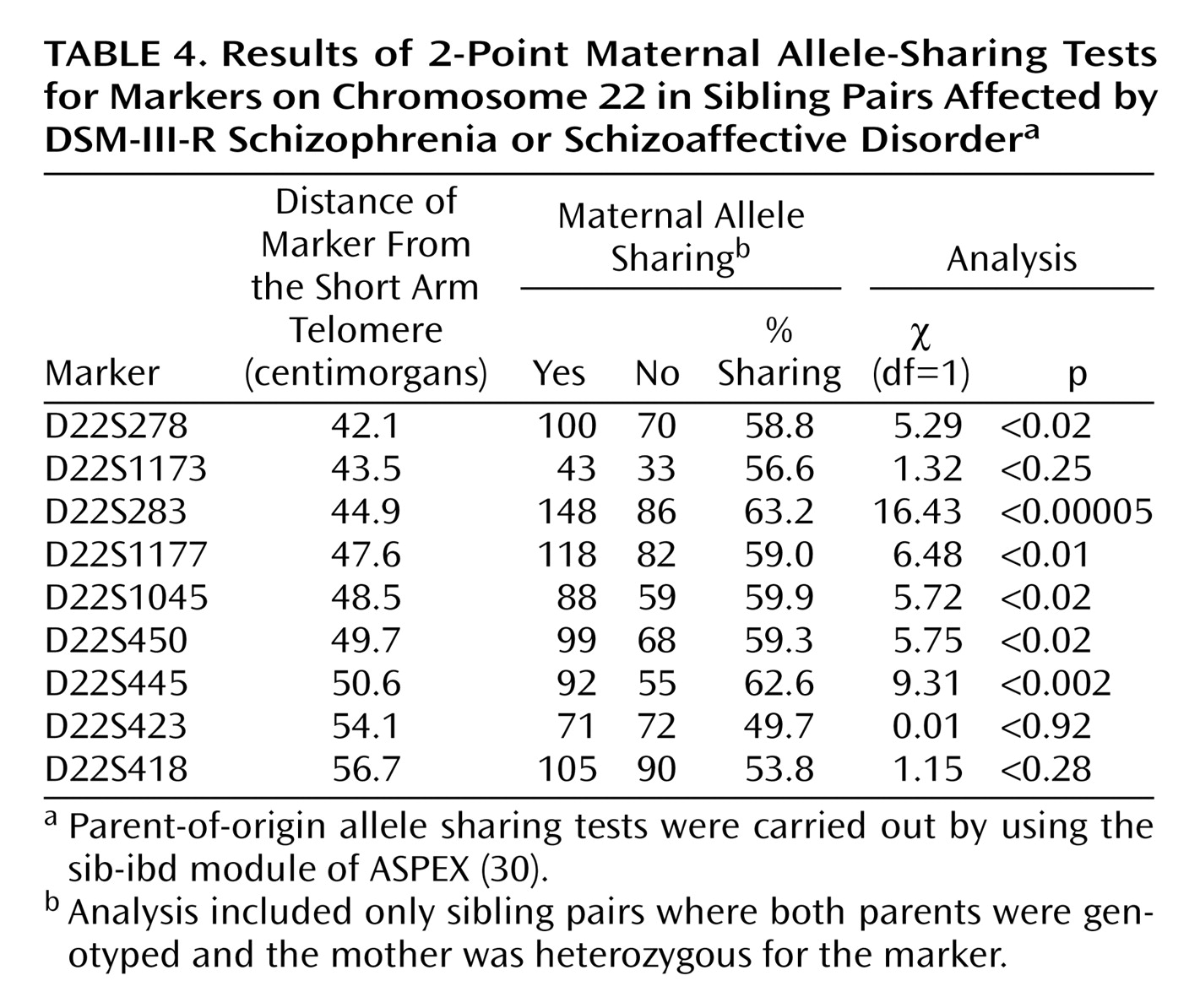
Because of the interest in a possible imprinting effect on chromosome 22q (Vallada and Collier
[34]), we investigated both maternal and paternal sharing of alleles among affected sibling pairs. Results of a chi-square test suggested a significant excess sharing of maternal alleles at locus D22S283 (χ
2=16.43, df=1, p=0.00005). The results of the 2-point maternal allele-sharing tests for all markers on chromosome 22 are shown in
Table 4.
Discussion
Genetic linkage studies of schizophrenia are currently in progress in independent laboratories worldwide. The limiting factor for most investigators has been the lack of availability of a large number of families for analysis. Thus, there have been many reports of suggestive linkage, but very few with significant findings. The present report contributes results from one of the largest complete genome-wide scans of schizophrenia to date. Although our findings when viewed on their own yield some evidence of linkage—for example, significant linkage in chromosome 10p14 and a suggestion of linkage in chromosome 2p14-q12—the most striking feature is the failure to confirm a number of earlier claims of positive findings.
We first consider the apparent evidence for linkage from our own studies. In an initial genome-wide scan
(16), chromosome 10p did not show a positive peak. However, others have previously reported 10p findings that suggest linkage to schizophrenia
(9,
35,
36), and Schwab et al.
(37) have presented data that phosphatidylinositol phosphate kinase on 10p is a candidate gene. However, like our findings for chromosome 2, the chromosome 10p findings are spread over a considerable distance. The finding in our present study (peak lod score of 3.60 at D10S189, 17cM from pter [the short-arm telomere]) is approximately 15–20 cM from the findings of Straub et al.
(9) (maximum heterogeneity lod score of 1.95 in a recessive model). It is 25–30 cM from the peak multipoint lod score of 2.13 obtained by Schwab et al.
(9), and at the short-arm end of the 60-cM band of positive nonparametric linkage scores of 1.70 to 3.36 from 10p13 to 10q23 reported by Faraone et al.
(36) in the European-American pedigrees of the National Institute of Mental Health (NIMH) data set. These scores are thus modest in magnitude and variable in location. Moreover, other systematic genome scans give no support to linkage on chromosome 10. Moises et al.
(38) in a two-stage approach in 70 pedigrees, Coon et al.
(39) in a large Palaoan pedigree, Brzustowicz et al.
(7) in 22 extended Canadian pedigrees, Williams et al.
(40) in their two-stage analysis of 196 affected sibling pairs, and Kaufmann et al.
(41) in African American pedigrees in the NIMH data all used markers with a resolution of 10 cM or less and failed to observe any positive peaks on 10p. Thus, there is little support in the literature for a conclusion that our multipoint lod score of 3.60 at D10S189 is evidence of a gene for schizophrenia on chromosome 10p14.
In our initial genome scan, we reported a peak lod score of 2.13 on chromosome 2p16.1 at D2S1337. In the current study, the expanded cohort had a peak lod score exceeding criteria for suggestive linkage as suggested by Lander and Kruglyak
(19) in the centromeric region of chromosome 2 near the marker D2S417 (102 cM from pter) and second peak in 2q12 at D2S160 (118 cM from pter). Other studies of families with schizophrenia have reported suggestive linkage for this region, although the associations have not reached a high level of statistical significance
(36,
38,
39,
42). Coon et al.
(39) reported a peak positive score on chromosome 2 at D2S441, which is approximately 98 cM from pter, and Faraone et al.
(36) reported a peak nonparametric linkage score of 2.41 on chromosome 2q12 at 104.5 cM from pter in a cohort of 43 nuclear families. Similarly, Levinson et al.
(42) reported a peak nonparametric linkage score of 2.01 at D2S410 located in 2q14.1 approximately 115 cM from pter in 45 pedigrees. In one of the earliest reported genome-wide scans for schizophrenia susceptibility genes, Moises et al.
(38) reported a nonparametric p value of 0.0001 at D2S135 located approximately 115 cM from pter in 2q12-2q14 in a group of Icelandic and international families. Brzustowicz et al.
(6,
7), while noting genome-wide evidence of significant linkage on chromosomes 1q and 13q, found a peak score of 2.42 at D2S1400, approximately 28 cM from pter. In a study of 198 affected sibling pairs, Williams et al.
(40) reported a wide peak on 2q at approximately 181 cM from pter, considerably distal to the peak in the present study. Kaufmann et al.
(41) reported modest peaks (nonparametric linkage scores <1.5) at 40 cM and 220 cM in African American pedigrees from the NIMH genetics consortium, with no evidence of linkage around the centromere. Thus, although all of these studies have reported findings on chromosome 2, the peaks are of modest magnitude and are widely dispersed along the chromosome. Only one of these findings
(39) comes within 4 cM of the present finding.
Chromosome 22q became a focus of attention after the report of Pulver et al.
(3) suggesting linkage to this chromosome in schizophrenia and descriptions of schizophrenia-like symptoms in patients with velocardiofacial syndrome, which arises from a deletion in chromosome 22q11
(43). This syndrome and its relationship to schizophrenia have been extensively studied
(44,
45). Some positive findings have been reported of linkage more distal to this region on 22q12
(3,
46,
47), and one report of a possible imprinting effect on this putative locus (linkage to maternally but not paternally transmitted alleles) in schizophrenia
(48) is consistent with our findings. Nevertheless, our weakly positive lod score for this region in our initial scan of 70 families did not increase further when the cohort was substantially enlarged. In addition, no evidence of linkage in this region was reported by Kaufmann et al.
(41), Faraone et al.
(36), Williams et al.
(40), or Brzustowicz et al.
(7). Similarly, the chromosome 3 and 12 suggestive linkages in the present study have not been reported to be of significance in any previous publications on schizophrenia.
Other regions of potential interest in our initial screen (chromosomes 1q, 4p, 5p, 11p, 16q) were not supported by further evidence of linkage in the fourfold larger cohort in the present scan. Moreover, a number of regions within which specific claims for linkage have been made by other investigators on 5q
(2), 6p 24-22 (e.g., reference
8), 13q
(5,
6), and 1q
(7,
14,
15) with small numbers of families have not shown linkage in the present group of 294 families. Two recent claims are of particular note. In 22 extended families, Brzustowicz et al.
(7) reported a heterogeneity lod score of 6.5 with an alpha (proportion of linked families) of 75% for a region in chromosome 1q211-q22 and claimed that the linkage result “should provide sufficient power to allow the positional cloning of the underlying susceptibility gene.” Leonard and colleagues
(48) reanalyzed data from the relatively small number of families from the NIMH consortium and obtained a lod score of 4.43 at 45.7 cM from the 15p telomere in a heterogeneity analysis with an alpha of 85%. If either of these claims were generalizable to a group of 294 families, we would expect to see lod scores substantially in excess of those reported in references
7 and
48. In fact, in our present study, we found no evidence of linkage to either of these regions. Although it is possible that some unusual skewness in the heterogeneity of our sample and those of others may explain our failure to replicate some previously reported findings, it is also possible that some of the reported linkages (or most) are false positive findings that will not lead to the location of a susceptibility gene.
Alternatively, it may be that the numbers of families with schizophrenia in all existing studies, including our own, are too small to replicate findings consistently. This argument has been supported by the simulated calculations of Goring and colleagues
(49) suggesting that if several genes of small effect are involved, as many as 3,000 sibling pairs would be needed to replicate results consistently. Thus, many positive regions with susceptibility genes could be missed even in a group of families as large as our current study group, and unusually large effects may be falsely seen with a much smaller number of families and may be falsely interpreted as evidence for the presence of susceptibility genes.
It is likely that the scope and complexity of linkage analysis combined with the idiosyncrasy of small study group sizes and a natural enthusiasm for positive findings has led investigators to conclusions that cannot be generalized to schizophrenia as a whole. We and others have not yet found a gene for susceptibility to schizophrenia or any other psychosis, although there is hope for the future, given the continued advances in technology. A candidate gene approach that takes into account the key underlying pathology present in patients with long-term schizophrenia (including brain structural, cognitive, and other physiological defects) may be a more fruitful direction. The possibility that linkage or association to a candidate gene may not be present owing to a lack of sequence variation needs to be considered, and the alternative possibility that the anomaly is epigenetic and related to variability in gene expression should be considered. Although the present results on chromosomes 2, 10, and 22 are intriguing and worth further pursuit, we acknowledge that they may be only chance findings awaiting replication and the detection of candidate genes within these regions.