Among these, the
KIAA0319 gene is a strong candidate, being supported by evidence in both association and functional studies. Significant associations have now been reported in at least three independent samples (
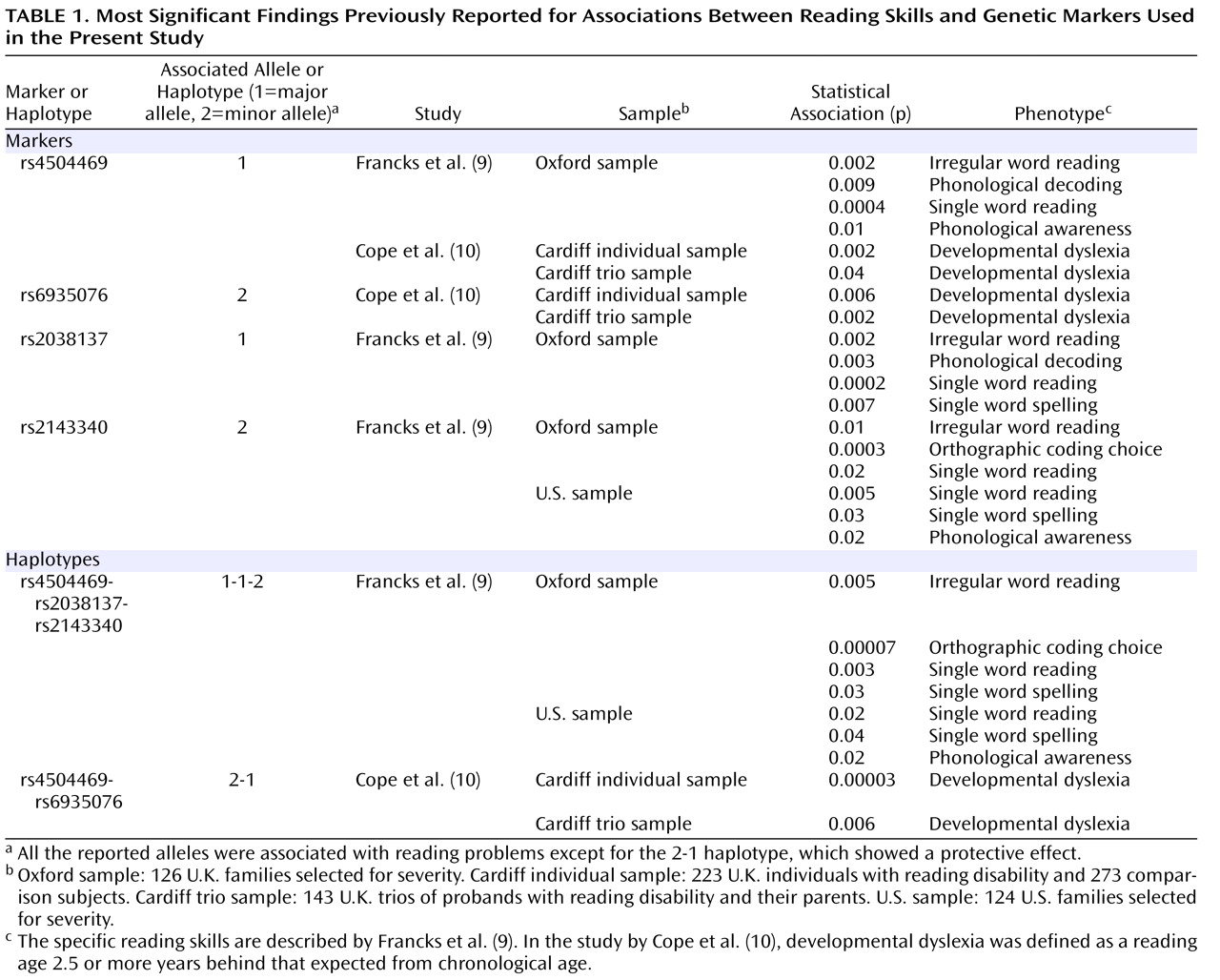
Table 1 ). Francks et al.
(9) identified a 77-kb region that showed association with different quantitative reading-related phenotypes in both a large sample of families from the United Kingdom and a sample of twin-based families from the Colorado Learning Disabilities Research Center. The region covered the first four
KIAA0319 exons, the entire
TTRAP gene, and the first exon of
THEM2 . The single-nucleotide polymorphisms (SNPs) rs4504469, rs2038137, and rs2143340 defined the major haplotypes present in the region, and the 1-1-2 haplotype (where 1 is the major allele and 2 is the minor allele), which is effectively tagged by the rs2143340 SNP, was significantly associated with reading disability in both samples. Cope et al.
(10) detected associations with markers rs4504469 and rs6935076, both located within
KIAA0319, in a completely independent sample of U.K. families by using a categorical definition of dyslexia. In particular, the 2-1 haplotype defined by these two markers was significantly associated with a protective effect, having a significantly higher frequency in good/normal readers. An association at the same locus was found in two other studies that, however, cannot be considered independent because they both derived their samples from the Colorado Learning Disabilities Research Center cohort
(15,
16) . Kaplan et al.
(15) reported association in a set of 104 families between a reading-related measure and the JA04 microsatellite, which is located in the first
KIAA0319 exon. Deffenbacher et al.
(16) performed association analysis in a set of 114 families selected for severity and detected evidence for association both at the
KIAA0319 locus and within the
DCDC2 gene, which lies about 200 kb distal to
KIAA0319 . A third study, which employed 153 families from the Colorado Learning Disabilities Research Center, showed an association with
DCDC2 but not with
KIAA0319 (11) . The different outcomes of these studies are most likely due to the different criteria employed for family selection and the small samples
(5) . Analysis conducted in a sample of German trios selected for spelling impairment showed association with the
DCDC2 gene
(12) .
No functional mutations have been identified yet for either of the two genes through resequencing of the exons and promoter regions
(9,
10,
12) . Harold et al.
(17) conducted a comprehensive study to compare the
DCDC2 and
KIAA0319 genes by genotyping the two U.K. samples previously used by Francks et al. (Oxford sample)
(9) and Cope et al. (Cardiff sample)
(10) for a common set of
DCDC2 and
KIAA0319 markers. None of the
DCDC2 markers showed any association, while several markers showed significant associations and identical allelic trends in both the Oxford and Cardiff samples. The most significant association was clustered around the first exon and putative regulatory sequences of
KIAA0319 in both samples, suggesting that the functional genetic variant might act at the level of gene expression. In agreement with this finding, we have shown through in vivo studies that the 1-1-2 haplotype is associated with a relative reduction of
KIAA0319 gene expression
(18) .
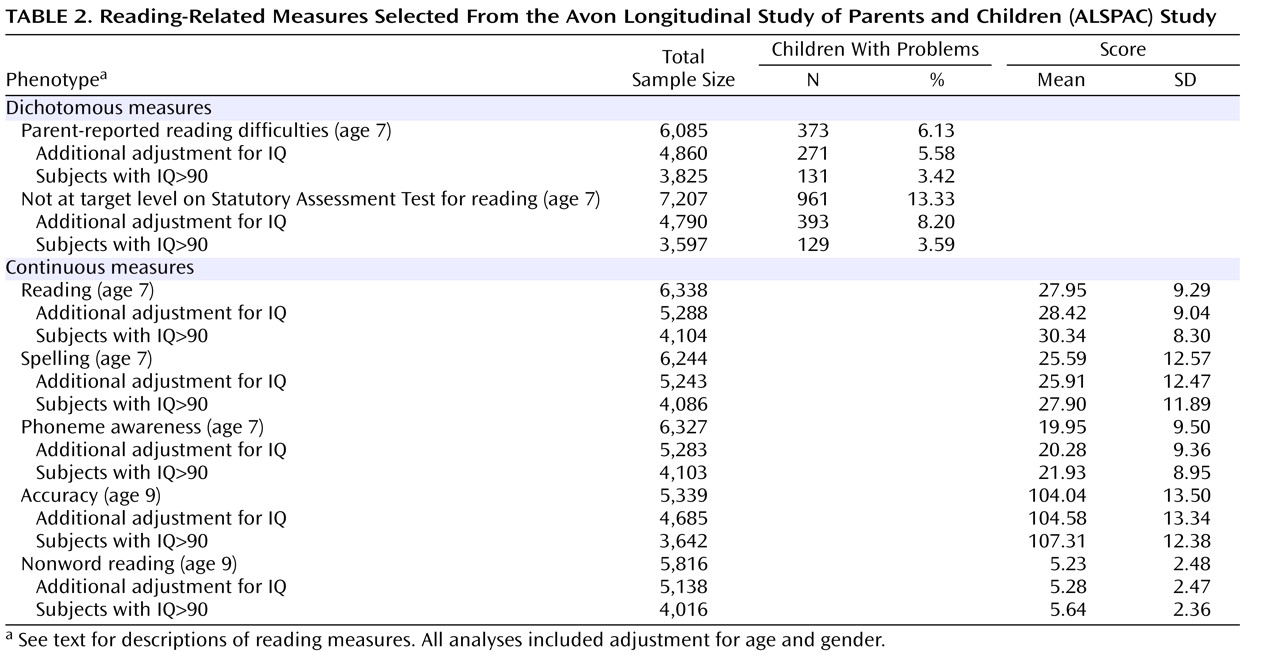
Reading disability could be considered to represent the lower tail of a normal distribution of reading ability observed in the population
(19) . It is possible that the quantitative trait loci underlying dyslexia susceptibility also influence variation in reading performance in the normal range. In the study described here, we aimed to test this hypothesis, asking whether the
KIAA0319 gene influences reading skills in the general population, rather than being specifically implicated in determining reading problems in restricted groups of individuals with dyslexia.
Results
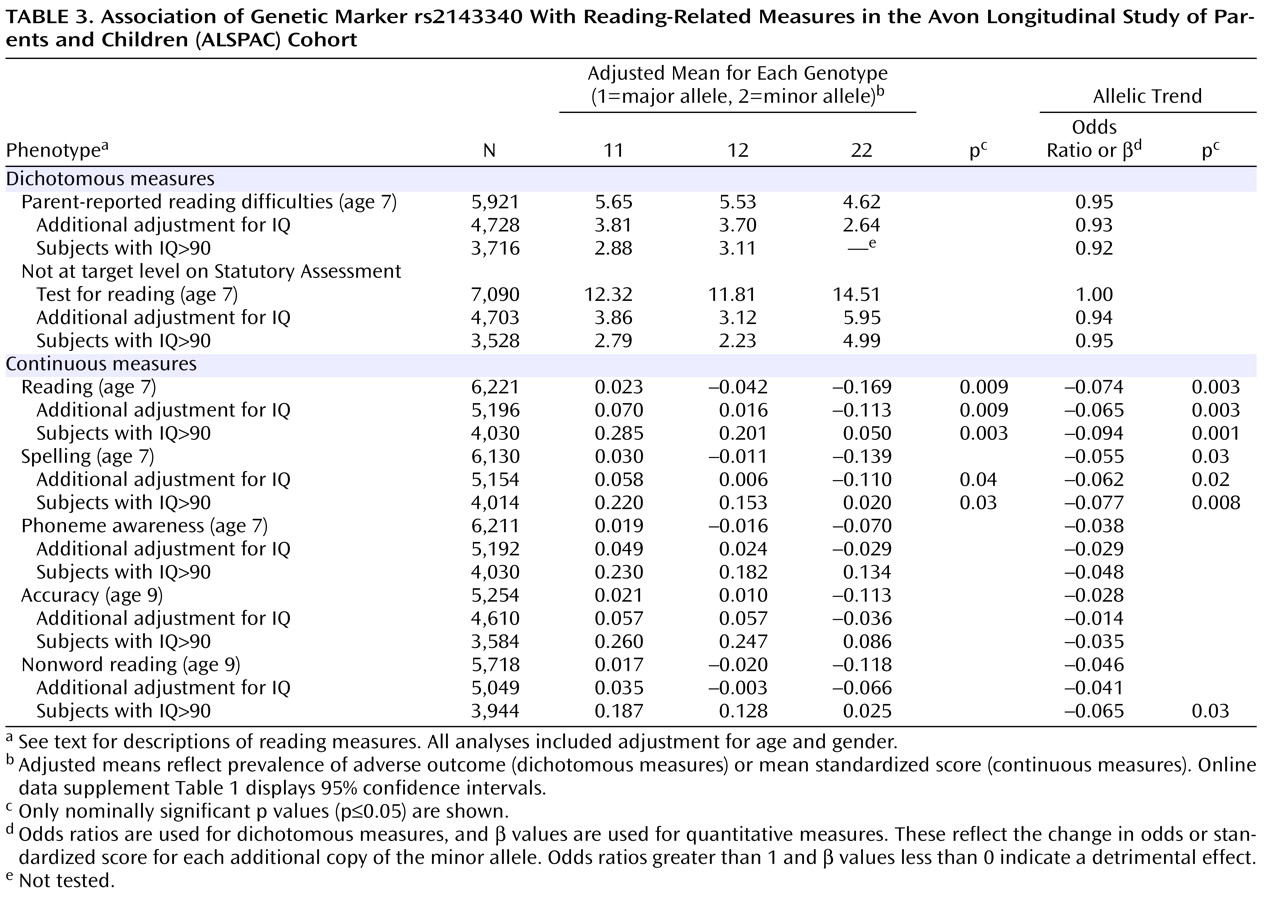
In the single-marker analysis, the SNP rs2143340 showed the most significant association. In particular, the minor allele was significantly associated with poor performance on the continuous measures of reading and spelling (
Table 3 ). The association with the reading test was nominally significant in the entire sample and became stronger after selection of individuals with high IQ. The association with spelling was of a lower magnitude but, similarly, increased in significance after control for IQ. The allelic trend model produced consistently stronger associations. Analysis with the other continuous measures showed a consistent trend of association between the minor allele of rs2143340 and poor performance (β<0,
Table 3 ). The other three markers showed only a few marginally significant associations (data supplement
Table 1 ). There was no particular trend of association for marker rs2038137, while rs4504469 showed mainly a trend of association between the major allele and poor performance in the reading-related measures, as previously reported
(9,
10) . The trend for rs6935076 was an association between the major allele and poor performance on the continuous measures, in disagreement with what was previously observed in the Cardiff sample, where a significant association with dyslexia was reported for the minor allele (
Table 1 )
(10,
17) . In our previous analysis of rs6935076 conducted in the Oxford sample, we did not detect any significant or consistent trend of association
(17) .
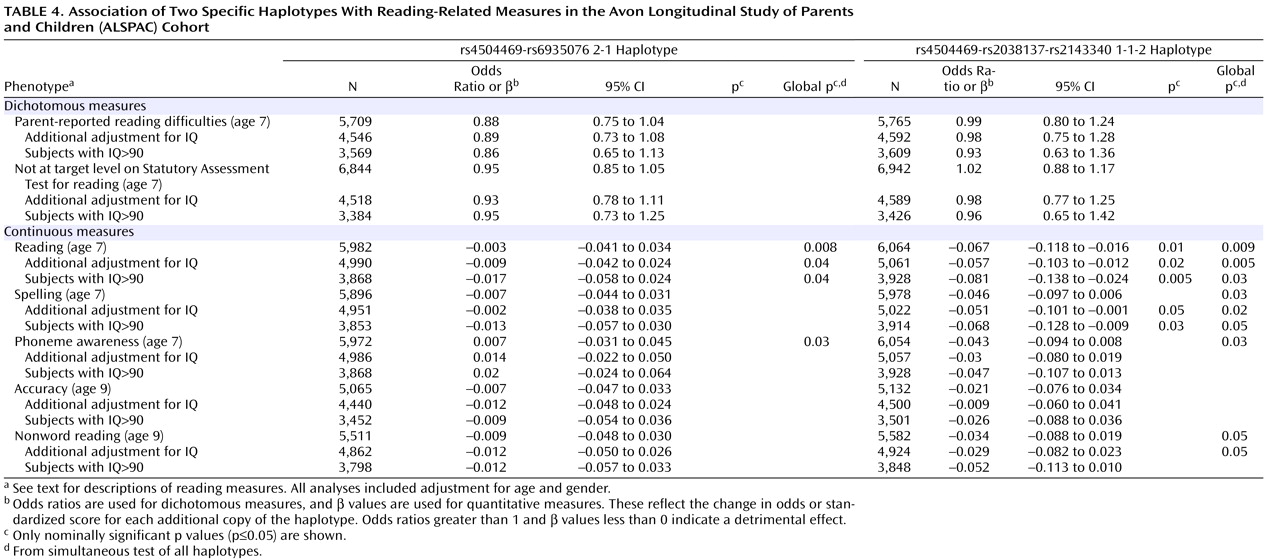
In the haplotype analysis, the 1-1-2 haplotype showed a nominally significant association with poor performance on the continuous measures of reading and spelling (
Table 4 ). A consistent trend of association between the 1-1-2 haplotype and poor performance was observed for all the continuous measures (β<0,
Table 4 ). The 2-1 haplotype did not show any significant associations. The test for “global” associations showed nominally significant associations for both the rs4504469-rs2038137-rs2143340 and the rs4504469-rs6935076 haplotypes (
Table 4 ). The three-marker haplotypes were associated with a wider range of measures, including reading, spelling, phoneme awareness, and nonword reading, while the two-marker haplotypes showed associations solely with the reading and phoneme tests.
Analysis of subgroups of individuals selected at the extremes of the distribution showed strengthening of the association of both rs2143340 and the 1-1-2 haplotype with the continuously scored reading phenotype in the 25th percentile selection of the entire sample (data supplement Table 2 and data supplement Table 3). Specifically, the analysis of the rs2143340 marker in this subgroup showed the strongest association observed in this study (allelic trend: p=0.0002). The same trend was not observed in more stringent percentile selections for the continuous reading measure. Other nominally significant associations were observed for other phenotypic subgroups, but overall, we did not observe a regular trend consistent with gradually increasing stringency in other phenotypic subgroups. An explanation for this lack of consistency could be the loss of power to detect the KIAA0319 effect in small samples. Similarly, we did not detect significant associations with the dichotomous measures when we tested small proportions of individuals with reading problems in a logistic regression framework. Most likely, when specific ascertainment criteria are not applied, quantitative analysis in a large sample reflecting all the phenotypic variation is more powerful than looking at smaller numbers of individuals at the extremes of the distribution.
Discussion
The results reported here are in agreement with our previous findings, which showed associations of the 1-1-2 haplotype and the minor allele of rs2143340 with poor reading performance
(9) . Moreover, all the associations we detected in the present study involving both the rs2143340 marker and the risk haplotype were strengthened after we controlled for IQ, and this was particularly true for the analysis of individuals with IQs above 90, supporting our hypothesis that IQ can be considered a confounder for the quantitative trait locus on chromosome 6p
(9) . Conversely, we did not observe any associations between the 2-1 haplotype and good reading performance, as previously suggested (
Table 1 )
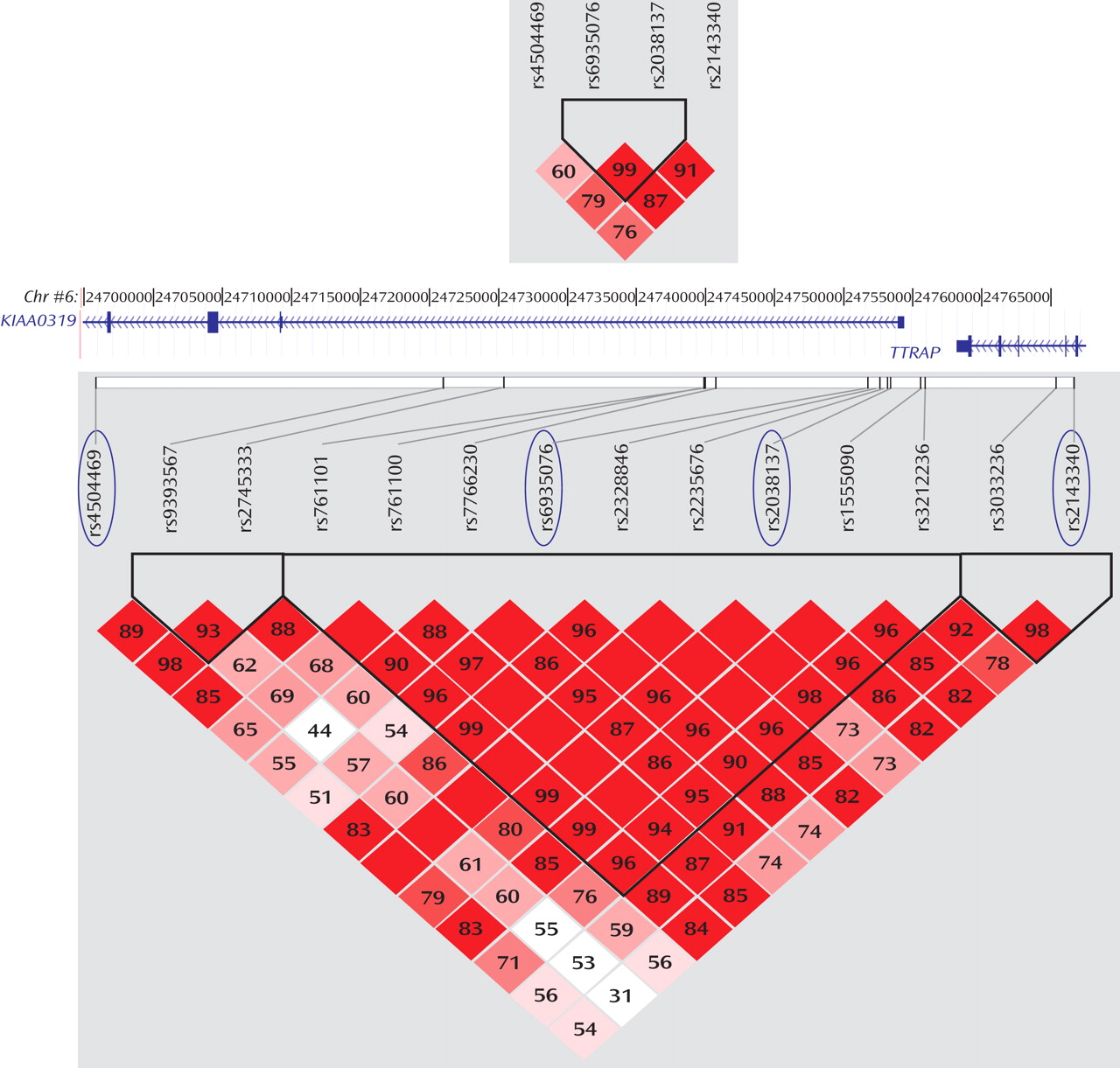
(10) . Nevertheless, the global test for the rs4504469-rs6935076 haplotypes showed some significant association, as would be expected owing to the correlation and physical proximity of the four SNPs (
Figure 1 ).
The allelic combination of markers rs4504469, rs2038137, and rs2143340 captured most of the genetic variation in the dyslexia-associated region by describing the most common haplotypes in our U.K. sample
(9) . We hypothesize that in the white European population, the 1-1-2 haplotype carries a functional mutation relevant to dyslexia. Association data in our previous
(9) and present study show that the haplotypic analysis does not detect any association stronger than what is observed in single-marker analysis. We conclude that the analysis of rs2143340 alone is sufficient and more powerful to detect the risk haplotype effect. This can be explained by the fact that rs2143340 efficiently tags the risk haplotype in the white European population, and missing genotypes in any of the three markers can contribute to loss of power in reconstructing haplotypes.
We have shown before that the 1-1-2 haplotype is associated with a reduction of
KIAA0319 gene expression in cell line models and that
KIAA0319 is required for neuronal migration during the development of the neocortex
(18) . We have hypothesized that a suboptimal level of the
KIAA0319 protein could be the cause of subtle cortical abnormalities that might lead to the development of dyslexia. A major question is, How is it possible that alteration in a gene involved in such a wide-ranging mechanism as neuronal migration can affect specific cognitive functions and not lead to a global impairment? Probably, the
KIAA0319 risk haplotype alone is not enough to have a significant impact on cognition, but when in the presence of particular combinations of multiple factors, it might specifically affect precise functions, such as reading abilities. Further studies are now required to identify such factors, of both genetic and environmental nature, and to test whether the
KIAA0319 gene might influence other phenotypes.
The present study shows that the effect of
KIAA0319 alleles conferring susceptibility to reading disability is not restricted to specific groups of dyslexic individuals. Sample selection and ascertainment criteria play a critical role in the outcome of association studies, which are normally based on samples of a few hundred probands. Individuals are usually recruited if their reading performance falls below a predetermined threshold defining dyslexia. There are not universally accepted criteria to fix such a threshold, and selection of different subgroups of individuals with reading disability might be at the basis of disparities in the outcomes of association studies
(5) . Negative association reports in previous studies
(11,
12) might be due to small samples that did not have enough power to detect the
KIAA0319 effect or in which the
KIAA0319 gene was not the major causative genetic factor. The same observation regarding sample size can be made for a recently published study that evaluated the influence of the
KIAA0319 gene on the reading skills of an unselected sample of 440 families in Australia
(29) . Associations were detected with both the rs2143340 marker and the 1-1-2 haplotype, but they cannot be regarded as identical allelic replications since they showed opposite trends; both the rs2143340 minor allele and the 1-1-2 haplotype were associated with better reading performance. The associations were only marginally significant and became further attenuated when the analysis was repeated only in individuals with at least 75% Anglo-Celtic ancestry (about 82% of the sample), suggesting that population admixture could also be a factor at the basis of this incongruous association. In support of this explanation, we observed that when we conducted the analysis on our entire sample, including individuals of nonwhite European ancestry, the associations were notably decreased (data not shown). For example, in the allelic trend tests, the strength of the association between rs2143340 and the continuous reading measure decreased from p=0.003 in the sample based only on individuals of white European ancestry to p=0.02 in the entire sample (gene-ethnicity interaction: p=0.04). This shows that a small number of individuals with different ancestry (about 4.6% of the sample analyzed) can significantly modify association outcomes, in agreement with previous reports
(30,
31) . The rs2143340 itself lies within the neighboring
TTRAP gene and probably does not have any functional role. More likely, it is in strong linkage disequilibrium in the white European population with a causal variant or variants expected to influence
KIAA0319 gene expression. As shown in
Figure 1, rs2143340 is in linkage disequilibrium with markers across
KIAA0319, including SNPs located at the promoter site, where gene expression regulatory regions are expected to reside. Conversely, the linkage disequilibrium between rs2143340 and
KIAA0319 promoter markers is lost in populations of Asian and African origin (www.hapmap.org; data supplement
Figure 1 ). If a functional mutation is indeed located at the
KIAA0319 promoter, it will then co-occur by chance more frequently with the major allele of rs2143340 in these other, non-European populations. This observation can explain the opposite trend of association detected in admixed samples when compared to samples of U.K. origin.
In conclusion, this study shows that the effect of a quantitative trait locus can be detected in an unselected sample, providing that the sample is large enough and reproduces the entire spectrum of a phenotypic distribution. These data not only provide further support for a role of the KIAA0319 gene in the development of dyslexia, but they also show that this gene influences reading ability in a wider context. Specifically, both the 1-1-2 haplotype and, in particular, the rs2143340 tagging SNP have been confirmed to be genetic risk factors for reading problems at the chromosome 6p quantitative trait locus in the U.K. population. These data suggest the need for functional studies both to identify the causal genetic variant responsible for the risk haplotype effect and to clarify the function of the KIAA0319 gene, in order to make it possible to dissect the biological pathway at the basis of the reading process and, more broadly, cognitive functions.