Stress responses and circadian rhythmicity are adaptations to environmental influences. Whereas stress responses are adaptations to unpredictable aspects of the environment, circadian rhythmicity is an adaptation to its predictable aspects, such as light-dark cycles (
2,
3). The hypothalamic-pituitary-adrenal axis and the extrahypothalamic brain stress system mediate behavioral and neuroendocrine reactions to stress in animals and humans (
2,
4). Both systems are under circadian control and thus provide an optimal adaptive response to environmental influences (
2,
5,
6). One important environmental risk factor for the development of alcohol use disorders is psychosocial stress. Exposure to early adverse life stress increases the risk of alcohol dependence in later life, and psychosocial stress in individuals abstinent from alcohol consumption increases alcohol craving and the propensity for relapse (
7–
9). In addition, stress-induced alcohol consumption can lead to disturbances in circadian rhythmicity, which might further exacerbate the use of alcohol (
10).
Although the interaction of stress response and circadian systems and their relevance to alcohol use have been documented at the behavioral and neuroendocrinological levels, the underlying integrative neurobiological mechanisms are not well established.
The molecular basis of circadian rhythmicity (circadian clock) involves a primary loop, with period 1, 2, and 3 (Per1, Per2, and Per3) and cryptochrome 1 and 2 (CRY1 and CRY2) proteins as transcriptional repressors and clock (homologue NPAS2 [neuronal period aryl hydrocarbon receptor nuclear translocator single-minded domain protein 2]) and BMAL1 (brain and muscle aryl hydrocarbon receptor nuclear translocator-like protein 1) proteins as transcriptional activators. This feedback cycle provides near-24-hour timing and drives the rhythmic expression of several clock-controlled and clock-modulated genes, which in turn mediate circadian rhythms in behavior and physiology. The circadian clock synchronizes and dictates the relative phasing of diverse internal physiological processes and molecular pathways. Clock genes also participate in reciprocal regulatory feedback, thereby rendering the circadian clock responsive to the internal environment of the body (
11).
There is increasing evidence that the circadian clock gene
Per1 is a target gene for glucocorticoids in mice (
12) and humans (
13) as well as stress-induced transcriptional activation in corticotropin releasing factor-expressing cells (
14). We hypothesized that
Per1 might be involved in the integration of stress response and circadian rhythmicity and aimed to explore this gene's relevance to alcohol drinking behavior.
Discussion
Gene-by-environment interactions account for substantial heterogeneity in response to environmental factors contributing to psychiatric disorders (
23). Circadian and stress-response systems are important physiologic mediators of environmental influences. In research on alcohol dependence, stress-related gene-by-environment interactions have been described (
1), and one prominent example relates to the finding that
CRHR1 moderates the effect of psychosocial stress on alcohol consumption in rodent animal models (
20,
24) as well as in humans (
25,
26). In the present article, we show that psychosocial stress moderates the effect of circadian rhythm gene
Per1 on excessive alcohol drinking. Mutant mice that lack a functional
Per1 gene, as a result of a genetic deletion of the PAS-binding domain, responded to repeated adverse stress exposure with enhanced alcohol consumption. This finding translates to our two human samples (one cohort of adolescents with a high adverse psychosocial load and one adult case-control sample of patients meeting DSM-IV criteria for alcohol dependence and comparison subjects, encompassing 2,457 individuals total).
We further examined underlying molecular mechanisms of the observed gene-by-environment interaction and conclude—from a series of in vitro experiments—that these associations might be caused by cortisol-induced transcriptional activation of
Per1, which appears to involve genotype-specific binding of the Snail1 transcription factor. Although it has been demonstrated that corticotropin-releasing factor is a primary mediator of extrahypothalamic behavioral stress response (
27), recent evidence suggests that stress-induced glucocorticoids can modulate the activity of the brain reward pathway and thus may account for individual vulnerability to the reinforcing effects of drugs of abuse (
9,
28).
The
Per1Brdm1-mutant mice did not differ in home-cage total alcohol consumption (for detailed information, see Zghoul et al. [
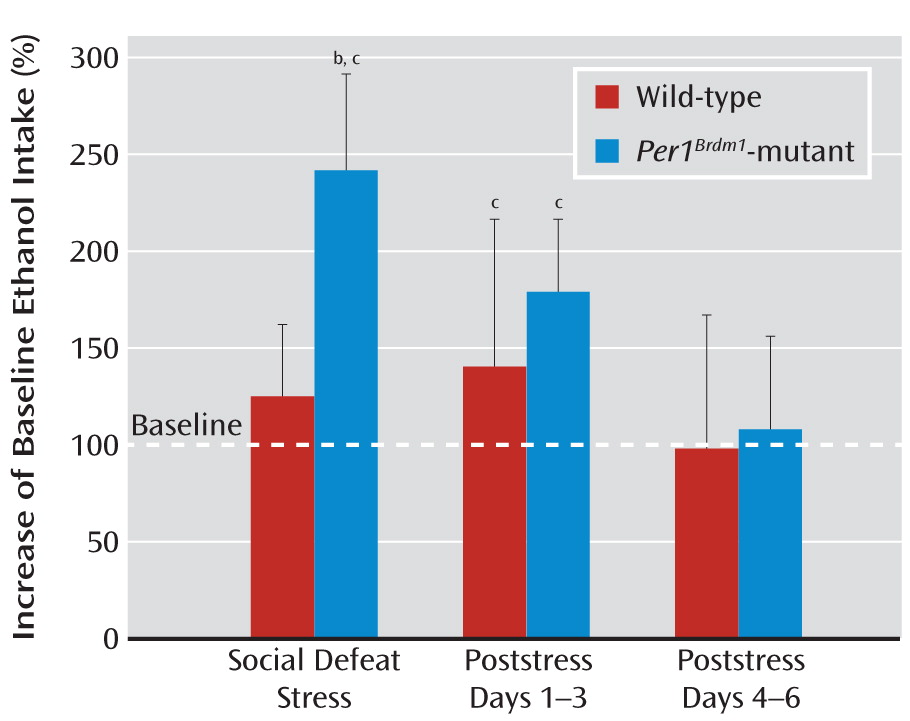
19]) but did exhibit a different noncircadian drinking pattern relative to their wild-type littermates. However, when exposed to repeated stress, the mutant mice showed more pronounced stress-induced alcohol intake than respective littermate comparison mice. Importantly, this effect was observed in three independent experiments applying different stressors (social defeat stress, swim stress, and foot shock stress). The most pronounced effects were observed with social defeat stress closely mimicking psychosocial stress in humans (
20).
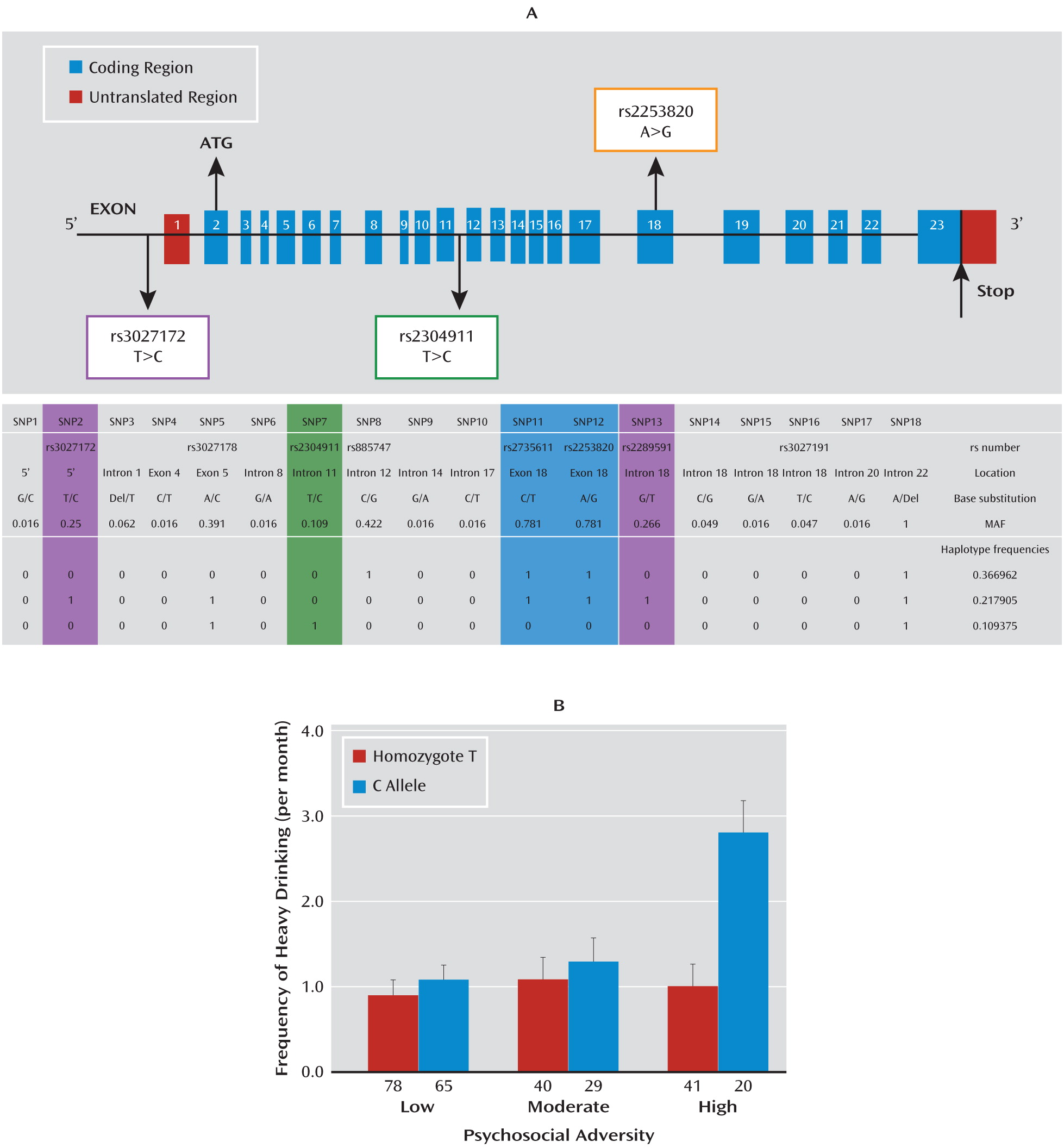
Our data further demonstrate that a genetic variation—SNP rs3027172—in the promoter region of the
hPer1 gene is associated with the effect of psychosocial stress on the frequency of heavy drinking in adolescents. In this age group, environmental influences are a more important determinant of alcohol consumption than they are in adulthood (
29). Problematic drinking behavior in adolescents with frequent episodes of heavy drinking is associated with coping motives, including coping with psychosocial stress (
30).
Frequent heavy drinking in adolescents in an attempt to cope with psychosocial stress is one entry mechanism into compulsive alcohol-dependent behavior (
31–
33), partly because of neuroplastic processes in the maturing brain, which confer a high vulnerability to drugs as well as alcohol (
33). In comparison, the association of the
hPer1 SNP rs3027172 with alcohol dependence in adults not only indicates the role of
Per1 as an entry mechanism into addictive behavior but also suggests its relevance for manifestation of alcohol dependence. The modest strength of the observed association may reflect the heterogeneity of the alcohol-dependent sample with respect to environmental exposure to adverse stress.
Cortisol is a main mediator of stress responding and serves as a master switch in the control of neuronal and network responses that underlie behavioral adaptations to stress (
2,
4). There is increasing evidence that
Per1 is a target gene for glucocorticoids, including cortisol in mice (
12) as well as in humans (
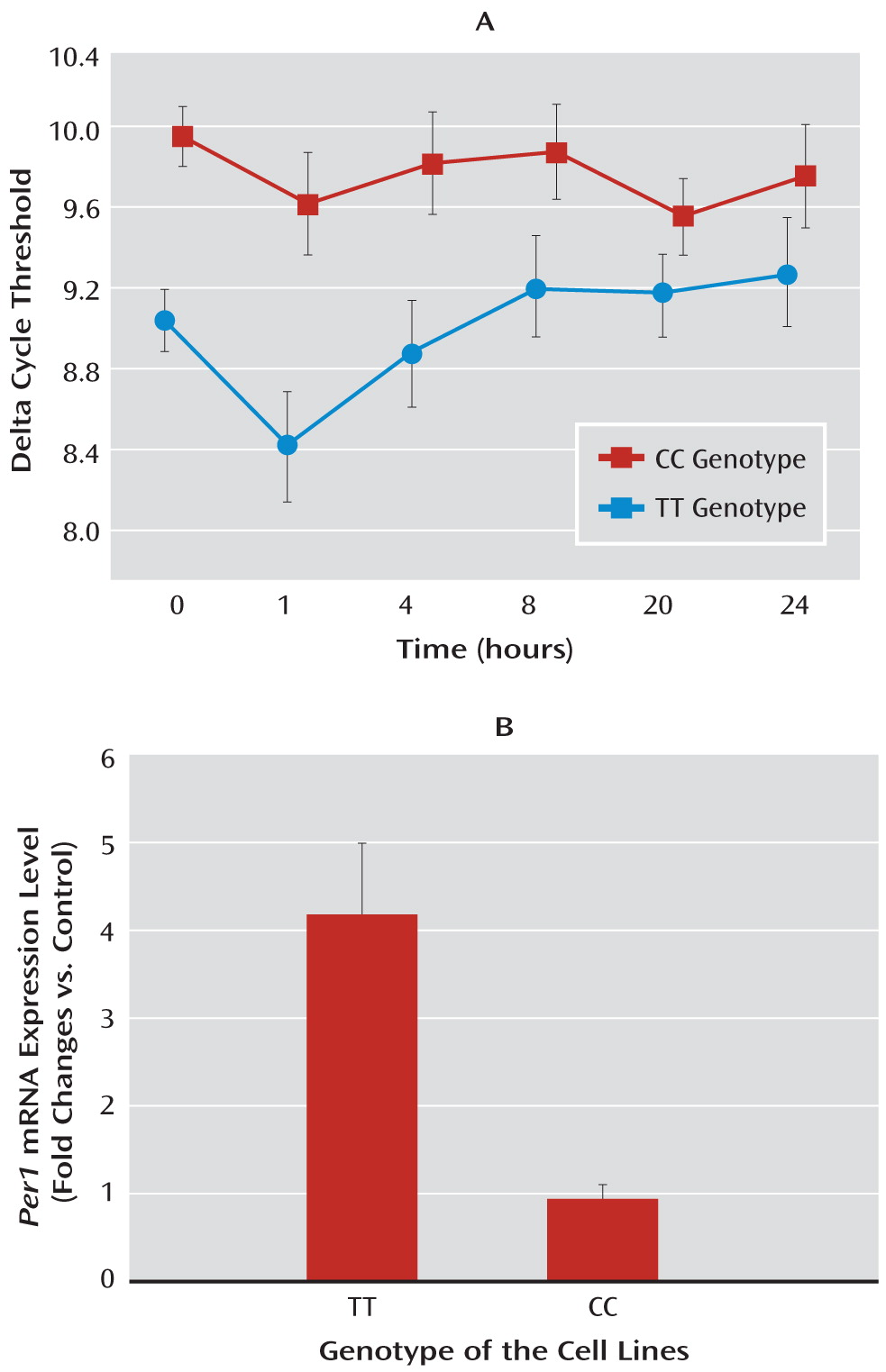
13). In line with these observations, we found significant cortisol-induced increase in the levels of
hPer 1 transcripts in human B-lymphoblastoid cell lines, which carry the protective genotype, but we did not find increased
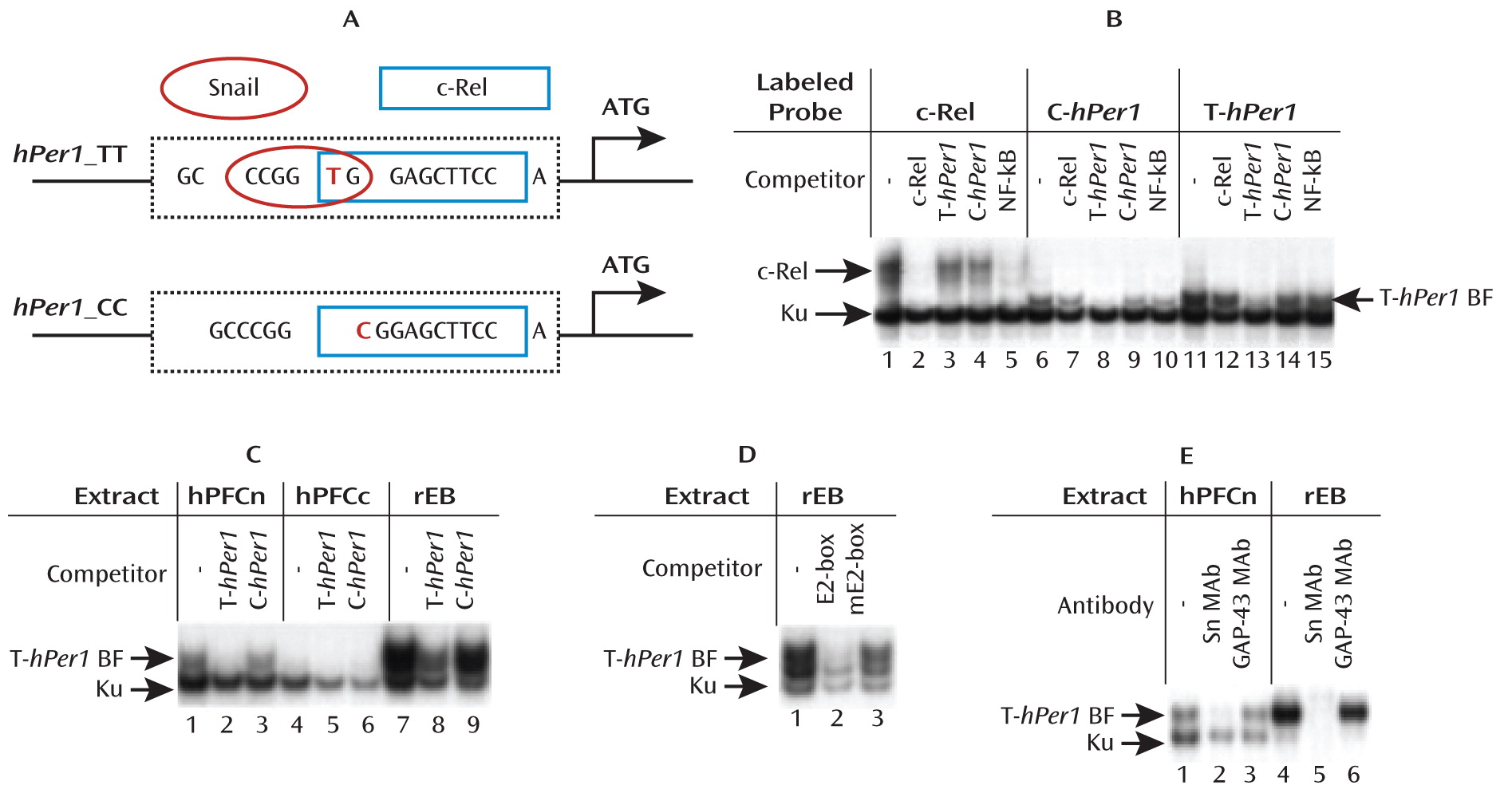
hPer 1 expression following cortisol exposure in cells carrying the risk genotype (rs3027172). Therefore, we hypothesize that there might be an altered response to stress-induced cortisol increase in carriers of the rs3027172 risk allele (C allele). Our electrophoretic mobility shift assay data further suggest that this altered stress response might be the result of a decreased binding affinity of the zinc finger protein Snail1 (
21,
22) to the rs3027172 C allele. This transcription factor is expressed in brain regions relevant to stress-induced drinking behavior, such as the prefrontal cortex, nucleus accumbens, and amygdala (
www.brain-map.org).
In conclusion, we provide evidence for an involvement of the
Per1 gene in psychosocial stress-induced alcohol drinking behavior. Based on the observation of increased stress-induced alcohol consumption in
Per1Brdm1-mutant mice, we demonstrated a gene-by-environment interaction whereby psychosocial stress moderates the association of risky alcohol drinking in adolescents with the
hPer1 promoter SNP rs3027172. This finding was further validated in an alcohol-dependent case-control sample. The rs3027172 genotype might be involved in cortisol-induced genotype-specific transcriptional activation of
hPer1, which is mediated in part by the transcription factor Snail1. Although the interpretation of gene-by-environment interaction in genetic association studies is complicated by the fact that environmental and genetic components are bi-directionally interconnected (i.e., the environment and the individual affect each other) (
25,
26), our in vitro and in vivo functional genetic analyses suggest that the effects of exposure to psychosocial stress on risky alcohol consumption are mediated in part by variation in the
Per1 gene.