Psychiatric disorders are common, and they are responsible for an enormous burden of suffering (
1,
2). Globally, approximately 18% of individuals suffer from mental illness every year (
3), and 44.7 million of those affected live in the United States (
4). Early detection and intervention for serious mental illness is associated with improved outcomes (
5–
8). However, few reliable predictors of risk or clinical outcomes have been identified. Given the substantial heritability of many psychiatric disorders (
9) and their polygenic architecture (
10), there is increasing interest in using quantitative measures of genetic risk for risk stratification (
11). Polygenic risk scores (PRSs) in particular are easy and inexpensive to generate and can be applied well before illness onset, making them a promising candidate for clinical integration (
12). A recent study investigating the clinical utility of PRSs for several common nonpsychiatric diseases (
13) found that these scores can identify a larger fraction of high-risk individuals than are identified by clinically validated monogenic mutations; the study authors call explicitly for evaluations of these scores in clinical settings.
To date, PRSs for neuropsychiatric disorders have primarily been evaluated in highly ascertained research samples. Typically, case subjects have obtained a diagnosis through lengthy clinician interviews, and control subjects have no psychiatric history (“clean” case and control samples). In order to bring PRSs to the clinic, however, they must first be demonstrated to have associations with diagnoses in real-world clinical settings, where data are often much messier. Among psychiatric disorders, schizophrenia is perhaps the best candidate for future clinical integration of PRS profiling, as it is highly heritable, has the best-performing PRS among psychiatric disorders in terms of proportion of phenotypic variance explained (7%) (
14), and may benefit from early detection and intervention (
5–
8). Accordingly, we selected the schizophrenia PRS for the present study, as it is the most viable test case for eventual clinical validation of a psychiatric PRS.
We recently established the PsycheMERGE consortium within the National Institutes of Health–funded Electronic Medical Records and Genomics (eMERGE) Network (
15,
16) to leverage electronic health record (EHR) data linked to genomic data to facilitate psychiatric genetic research (
17). In this first report from PsycheMERGE, we evaluate the performance of a schizophrenia PRS generated from summary statistics published by the Psychiatric Genomics Consortium (PGC) (
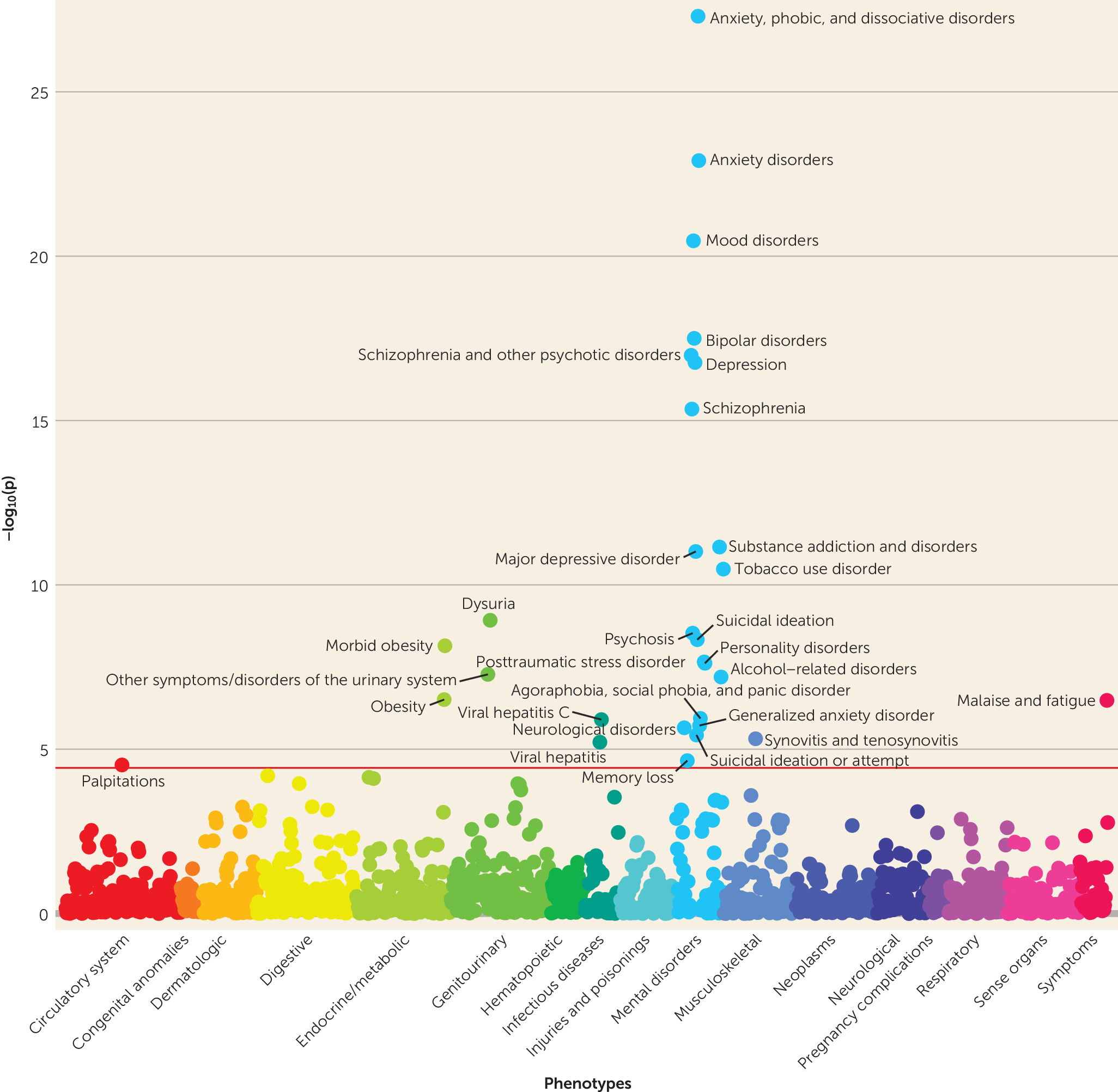
14) using EHR data on more than 100,000 patients from four large health care systems (Geisinger Health System, Mount Sinai Health System, Partners HealthCare System, and Vanderbilt University Medical Center). We assessed the relative and absolute risk for schizophrenia among individuals at the highest level of genetic risk and considered the clinical utility of the PRS for risk stratification. We also examined pleiotropic effects of the schizophrenia PRS with real-world clinical data by conducting a phenome-wide association study (PheWAS) of 1,359 disease categories.
Finally, we conducted follow-up analyses to characterize the nature of the pleiotropic effects of the schizophrenia PRS. Cross-phenotype associations of polygenic liability to schizophrenia may occur in at least two scenarios (
18). In the first, “biological pleiotropy,” the PRS contributes independently to multiple phenotypes. In the second, “mediated pleiotropy,” the PRS increases liability to a second disorder that occurs as a consequence of schizophrenia itself. For example, an association between schizophrenia polygenic risk and diabetes could occur because individuals diagnosed with schizophrenia are more likely both to have elevated schizophrenia PRS and to receive prescriptions for antipsychotic medications, which may result in weight gain and increased liability to diabetes. In this case, the observed relationship between schizophrenia risk and diabetes is mediated by the use of antipsychotic medication. Such scenarios may be difficult to completely disentangle. However, here we use individual-level EHR data to determine whether associations with genetic risk for schizophrenia persist after conditioning on a clinical diagnosis of schizophrenia, related psychosis, or prescription of antipsychotic medication.
Discussion
We investigated the impact of genetic risk for schizophrenia across the medical phenome in 106,160 patients from four large U.S. health care systems. Several findings from our analyses are noteworthy. First, externally derived PRSs for schizophrenia robustly detected risk for diagnosis of schizophrenia (phecode 295.1) in real-world health care settings (p values <4.48×10
−16). The effect sizes (see
Table 2) were similar to those observed for corresponding PRSs for atrial fibrillation, type 2 diabetes, inflammatory bowel disease, and many common cancers (
13,
31). Second, we leveraged the phenome-wide data available in EHRs to conduct the first psychiatric PRS PheWAS in multiple U.S. health care systems, revealing a range of pleiotropic relationships.
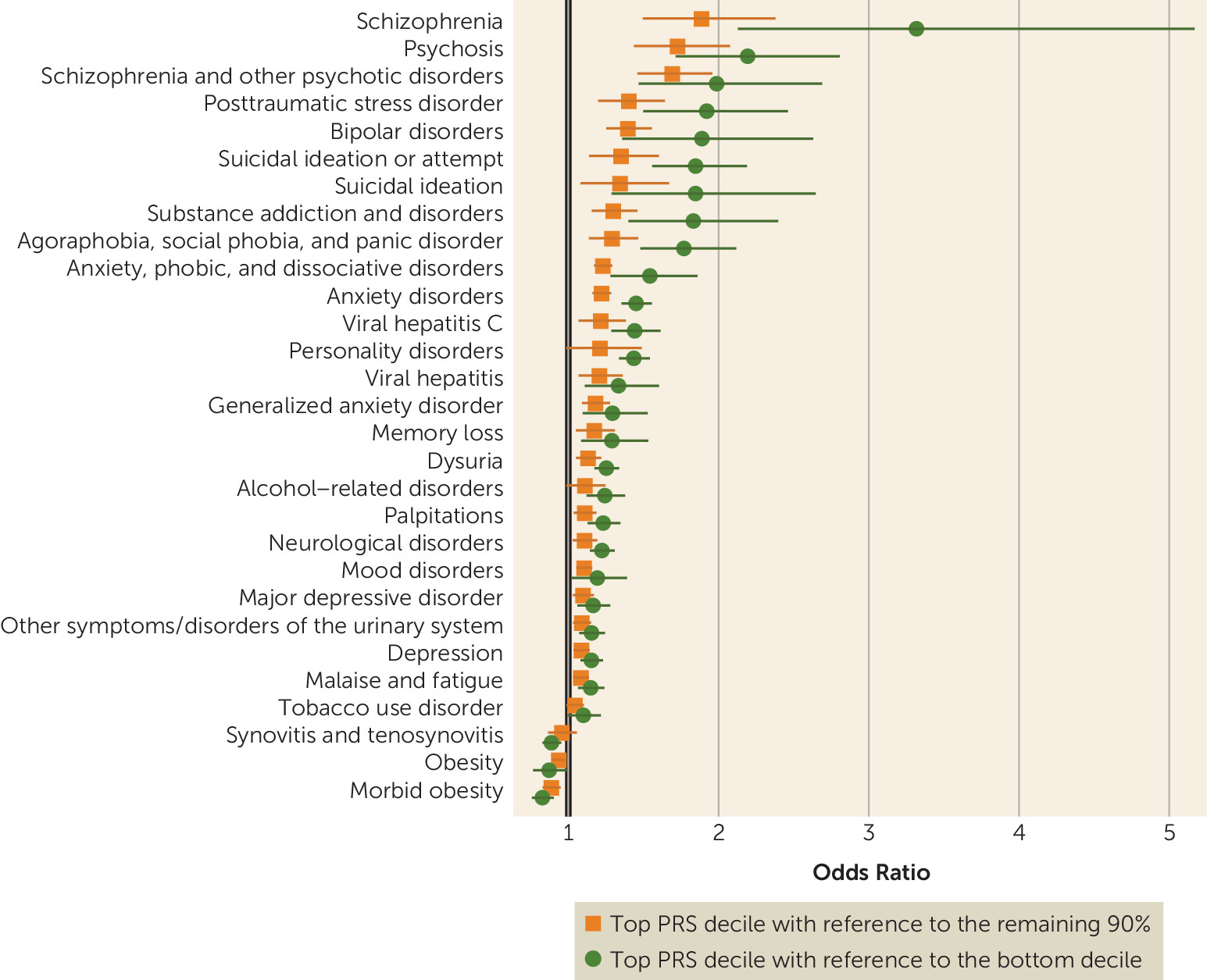
While we observed strong associations with schizophrenia, the effect sizes were more modest than those reported in schizophrenia case-control cohorts ascertained for research purposes. For example, in the original report by the PGC from which the risk scores were derived, depending on the sample, individuals in the top decile of schizophrenia PRS relative to the bottom decile had 7.8-fold to 20.3-fold greater odds of schizophrenia (
14), whereas we observed odds ratios of 3.3 and 4.6, depending on the PRS method (see
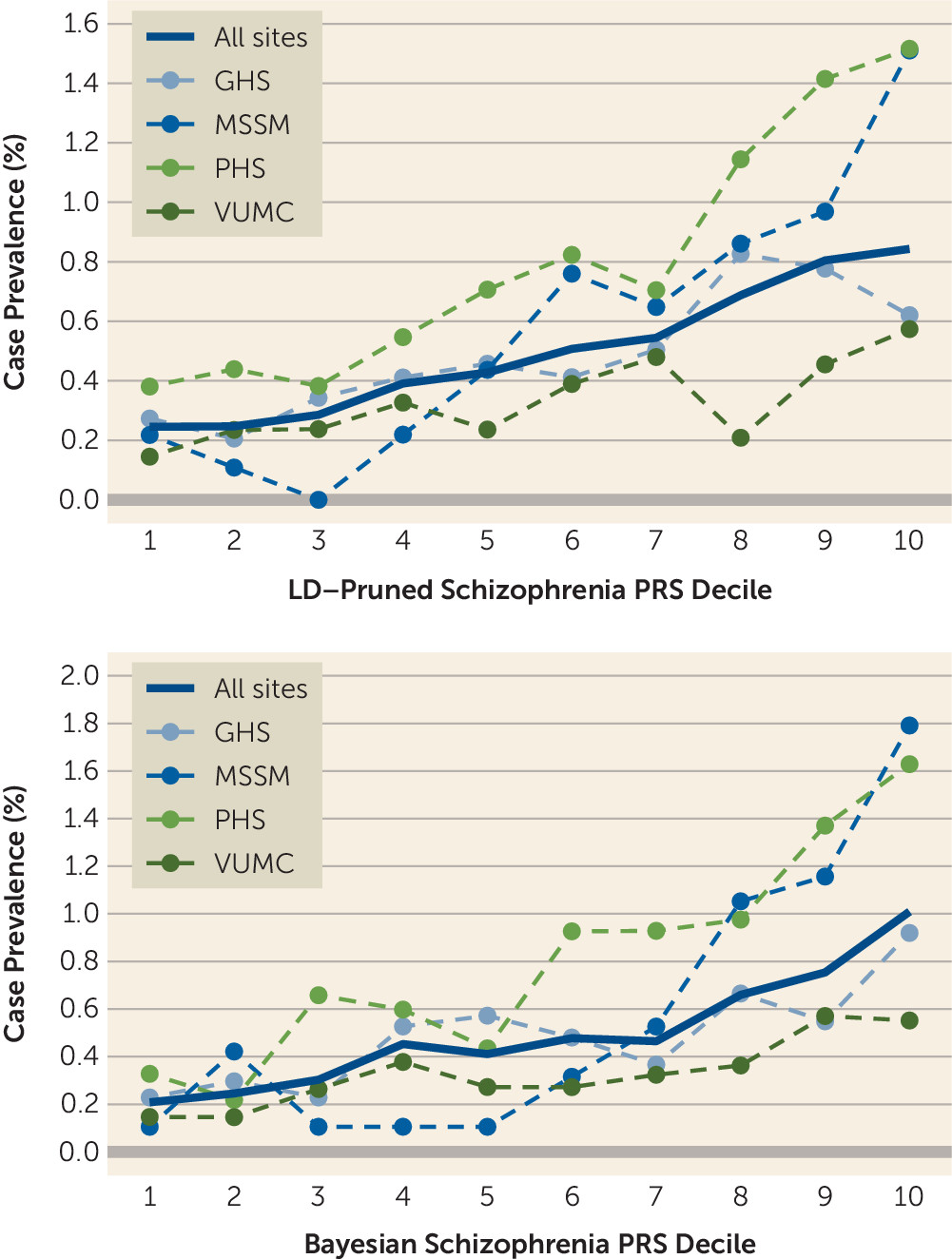
Table 2). There are several potential reasons for this discrepancy. First, case subjects in the PGC meta-analysis met relatively stringent criteria based on clinical interviews by trained research personnel, and control ascertainment often included screening for history of psychiatric or neurological disorders. This approach, typical for research samples, maximizes power for genetic discovery by extreme sampling from the tails of the genetic liability distribution. In contrast, our study used passively collected clinical data—participants were not asked to do anything outside of routine clinical care, thus reducing barriers to participation—and we did not set a clinical symptom threshold for control subjects (other than that they could not be case subjects). This approach more closely approximates an epidemiological design, similar to health registry–based studies in European countries. Thus, although the effect size we observed is likely attenuated as a result of some degree of misclassification, it may better reflect results that would be seen in real-world clinical settings where PRSs were applied to a broad health care population, with little a priori knowledge of clinical symptoms. In addition, we did not restrict the age range of case and control subjects, which may have further reduced the apparent effect size of the schizophrenia PRS (some individuals in our sample who have not yet reached the age of illness onset may have been misclassified as control subjects).
Although the PRS effects we observed were not large enough on their own to stratify risk in a clinical setting (i.e., to discriminate between cases and controls on an individual level with high accuracy), they are comparable to those of risk factors in established risk calculators. For example, two well-established coronary artery disease risk factors, smoking and diabetes, were estimated in the Framingham Heart Study to have hazard ratios ≤2.0 (
32)—similar to the observed risk for the top schizophrenia PRS decile here. In light of this, we speculate that incorporating genetic risk could have an impact in psychiatry, especially as enhanced performance may be possible through a variety of means. For example, we implemented two PRS methods, a standard LD-pruning approach and a newer Bayesian one, to evaluate the robustness and consistency of our results. While the differences in results were not large, the Bayesian method produced larger effect estimates overall, including for schizophrenia (see
Table 2). These findings support the use of newer risk scoring methods that can incorporate more genetic variants by directly modeling LD structure. The precision of PRSs may also increase through larger discovery sample sizes (
12) and with refinement of EHR-based case definitions.
Nonetheless, it remains to be seen whether combining PRS risk estimates with other clinical predictors can meaningfully contribute to individualized risk assessment in psychiatry. As expected, the areas under the receiver operating curve—a common metric used to evaluate predictive performance—for PRS alone were modest (0.60–0.71 across sites; see Table S6 in the
online supplement), although they were similar to those observed for schizophrenia PRSs in research samples (0.59–0.81) (
14), as well as similarly computed PRSs for other complex diseases, including type 2 diabetes (0.70), breast cancer (0.66), and inflammatory bowel disease (0.60) (
13). A remaining challenge for all risk stratification efforts in low-prevalence diseases (such as schizophrenia) is that even at high risk thresholds (e.g., the top 10% of a PRS), most individuals are not affected, limiting the utility of stratification for clinical practice. It may be that adequate precision will only be achieved through incorporation of many different measures of risk (e.g., genetic and nongenetic factors).
Schizophrenia PRSs were also associated with broader effects on mental health, including higher risks for anxiety, mood, substance use, personality, and neurological disorders, as well as memory loss and suicidal behavior. Anxiety, mood, and substance use disorders have all previously been linked to genetic risk for schizophrenia (
9,
33–
35), and our results confirm in a clinical setting that these disorders share genetic risk. Certain personality disorders have also been linked to genetic liability for schizophrenia (
36,
37) (e.g., schizotypal or schizoid), and there is some evidence that personality dimensions in adolescence predict future psychopathology, including schizophrenia (
38). Similarly, family history of schizophrenia has been associated with suicidal behavior (
39). However, results from our sensitivity analyses suggest that the relationships between schizophrenia and neurological disorders, personality disorders, suicidal behavior, and memory loss may be consequences of a schizophrenia diagnosis rather than due to shared genetic risk (see Figure S2 in the
online supplement).
Genetic liability for schizophrenia was associated with many nonpsychiatric syndromes as well, including obesity, urinary syndromes, viral hepatitis, synovitis and tenosynovitis, and malaise and fatigue. Intriguingly, obesity and morbid obesity were significantly negatively associated with schizophrenia PRSs (see Table S1 in the
online supplement). This is somewhat surprising given the known phenotypic correlation between schizophrenia and obesity (
40). Nonetheless, three previous reports found significant inverse genetic correlations between body mass index and schizophrenia (
41–
43), while a fourth reported an inverse but nonsignificant relationship (
44). This may suggest that elevated rates of obesity among patients with schizophrenia may be a consequence of the disease, potentially due to antipsychotic use or poor support for proper nutrition. We also found an inverse association between genetic liability for schizophrenia and diabetes, but only in sensitivity analyses controlling for a schizophrenia diagnosis or antipsychotic medication history. It may be that this negative genetic correlation was attenuated in the primary analysis (i.e., including patients with schizophrenia and antipsychotic medication history with no statistical control) because of diabetes-promoting effects of antipsychotic medications within the same individuals who were at high genetic risk for schizophrenia (
40). In general, pleiotropic effects may have implications for risk communication if PRS testing is deployed in clinical settings in the future.
Our results should be interpreted in light of several limitations. First, because of small numbers of patients of other ancestries, our analyses were restricted to patients of European descent, and the generalizability to individuals of non-European ancestry remains to be determined. Second, our phenotype definitions relied on very simple rules and disregarded many variables of potential importance, including medical history of related disorders, setting of diagnosis (i.e., inpatient or outpatient; physician specialty), and treatment for the disease of interest. This was by design in order to mimic a real-world clinical population in which PRSs may be implemented for clinical decision support; however, the approach is sensitive to misclassifications that occur in a clinical setting. Future work refining case and control definitions using natural language processing algorithms may improve the predictive performance of PRSs and other risk factors for clinically derived phenotypes (
45,
46). Third, our results varied to some degree between sites (see Tables S3 and S4 in the
online supplement), perhaps most notably for schizophrenia, suggesting that demographic and disease distributions in any given health care system will influence penetrance and pleiotropy. However, we tested for between-site heterogeneity for schizophrenia (phecode 295.1), and although this test has relatively low power, it showed no evidence of significant heterogeneity (p values >0.45). Relatedly, disease prevalence was often lower in the overall health care system relative to the participants enrolled in the biobanks (a subset of those patients) (see Table S7 in the
online supplement). In general, case prevalence in the biobanks was more representative of population-level prevalence than it was in the health care systems, suggesting that the discrepancies may be due to biobank patients generally having a longer duration of EHR follow-up and therefore more opportunity to receive a diagnosis than patients in the overall health care system (see Table S7). Finally, although our analyses comprise the largest test of a schizophrenia PRS in EHR data to date, additional phenotypes may show significant association in future larger-scale PheWASs.