Neuregulin-1 and its receptor ErbB4 have been implicated in the pathogenesis of schizophrenia, and this association appears to involve dysregulated splicing of ErbB4 transcripts (
1,
2). For example, transcript levels of some ErbB4 splicing variants are higher in the dorsolateral prefrontal cortex of schizophrenia subjects (
3–
5), whereas total ErbB4 mRNA levels are unaltered (
3,
4). Furthermore, the elevated ErbB4 splicing variants are associated with the intronic single-nucleotide polymorphisms (SNPs) at the ErbB4 locus that are linked to schizophrenia (
4,
6). Finally, levels of myocardial infarction associated transcript (MIAT), a noncoding RNA that regulates the splicing of ErbB4 transcripts, are altered in schizophrenia (
7). In concert, these findings support a role for dysregulated ErbB4 splicing in the pathogenesis of schizophrenia.
Alternative splicing of ErbB4 pre-mRNA results in four ErbB4 variants, each with different functional effects (
8). Splicing at the juxtamembrane (JM) locus produces the minor JM-a and major JM-b variants based on the inclusion of exon 16 or exon 15b, respectively (
9). The inclusion of exon 16 renders the JM-a isoform susceptible to proteolytic cleavage at the juxtamembrane domain (
10–
12). Splicing at the cytoplasmic (CYT) locus produces the minor CYT-1 and major CYT-2 variants based on the inclusion or exclusion of exon 26, respectively. Both the CYT-1 and CYT-2 isoforms couple to the MAPK signaling pathway, whereas only the CYT-1 isoform activates the phosphoinositide 3-kinase (PI3-K)-Akt pathway (
13). Thus, although alternative splicing clearly influences ErbB4 signaling, the effects of dysregulated ErbB4 splicing on dorsolateral prefrontal cortex function in schizophrenia are unknown.
In mouse prefrontal cortex, ErbB4 is preferentially expressed in parvalbumin-positive interneurons (
14), where it regulates the formation of excitatory inputs to, and hence the activity of, these cells (
15,
16). The activity of cortical parvalbumin interneurons is thought to be reduced in schizophrenia since the expression of two activity-dependent transcripts (
17–
19), parvalbumin and glutamic acid decarboxylase (GAD67), are lower in parvalbumin interneurons in schizophrenia subjects (
20–
23). Consistent with these findings, protein levels of parvalbumin and GAD67 are also lower in parvalbumin axon terminals in schizophrenia (
24,
25). The idea that these alterations in parvalbumin neurons could be attributed to disturbed ErbB4 signaling is supported by findings that deletion of ErbB4 in parvalbumin interneurons in mice results in lower parvalbumin interneuron activity accompanied by lower parvalbumin and GAD67 levels (
15).
Similar to how it appears in mouse prefrontal cortex, ErbB4 is expressed in virtually all parvalbumin interneurons in primate dorsolateral prefrontal cortex (
26). Parvalbumin interneurons are heavily enriched in layer 4 of the dorsolateral prefrontal cortex, and lower parvalbumin mRNA levels are restricted to dorsolateral prefrontal cortex layers 3 and 4 in schizophrenia subjects (
20). ErbB4 is also expressed in all calretinin-positive interneurons, which are preferentially localized to primate dorsolateral prefrontal cortex layer 2. Interestingly, calretinin mRNA levels are not altered in schizophrenia (
20,
27), suggesting that calretinin interneurons are relatively intact in the disease.
Therefore, in this study we sought to determine whether dysregulated ErbB4 splicing is associated selectively with parvalbumin interneurons, and not with calretinin interneurons, in schizophrenia. First, we hypothesized that in schizophrenia the levels of ErbB4 splicing variants are altered in dorsolateral prefrontal cortex layer 4, where parvalbumin interneurons are heavily enriched, and not in layer 2, where calretinin interneurons are highly localized. Second, we hypothesized that dysregulated ErbB4 splicing in schizophrenia is associated with lower parvalbumin mRNA levels in dorsolateral prefrontal cortex layer 4. Finally, we hypothesized that the altered levels of MIAT and/or the schizophrenia-associated intronic SNPs at the ErbB4 locus contribute to dysregulated ErbB4 splicing in schizophrenia.
Method
Human Subjects
Brain specimens were obtained during routine autopsies conducted at the Allegheny County Office of the Medical Examiner (Pittsburgh) after consent was obtained from the next of kin. Consensus DSM-IV diagnoses for each subject were made by an independent team of clinicians using structured interviews with family members and review of medical records (
23). The absence of a psychiatric diagnosis was confirmed in healthy comparison subjects by using the same approach.
To control for experimental variance and reduce biological variance, each subject with schizophrenia or schizoaffective disorder was matched with one healthy comparison subject for sex and as closely as possible for age. For the initial analysis of ErbB4 variant expression in gray matter, 62 pairs of subjects were used. For the layer-specific analysis of ErbB4 variant expression, a subset of these pairs (N=39) were used because of limitations in the availability of tissue sections for laser microdissection (see Table S1 in the
data supplement accompanying the online version of this article). The mean age, postmortem interval, RNA integrity number (Agilent Bioanalyzer, Agilent Technologies, Santa Clara, Calif.), and tissue storage time did not differ between subject groups for either the 62 or 39 subject pairs (
Table 1). Although brain pH significantly differed between subject groups (t=2.68, df=61, p=0.009 for 62 pairs; t=3.12, df=38, p=0.003 for 39 pairs), the mean differences were small (0.1–0.2 pH units). Each subject had an RNA integrity number of at least 7.0, indicating an excellent quality of total RNA.
Total and Layer-Specific RNA Extraction
Tissue was collected from dorsolateral prefrontal cortex area 9, and total RNA samples were prepared as described previously (
23) (see Supplementary Methods section in the online
data supplement). Fresh-frozen sections (12 µm) from area 9 were mounted on microdissection slides (Leica Microsystems, Bannockburn, Ill.), fixed in ethanol/sodium acetate, stained in 0.5% thionin, and dehydrated with ethanol as previously described (
28). The boundaries of layers 2 and 4 were determined on the basis of the size and packing density of the stained cells. A Leica microdissection system (LMD 6500, Leica Microsystems, Wetzlar, Germany) was used to collect ∼3×10
6 µm
2 of tissue from each of layers 2 and 4 per subject from the same section. The collected tissue was lysed by vortexing for 30 seconds in 200 μl of RLTplus buffer (Qiagen, Valencia, Calif.). RNA was extracted by using the RNeasy Plus Micro Kit (Qiagen, Germantown, Md.), and complementary DNA was synthesized by using the qScript cDNA SuperMix (Quanta Bioscience, Gaithersburg, Md.) as previously described (
28).
Quantitative Polymerase Chain Reaction (qPCR)
Primer sets were designed to quantify transcript levels for parvalbumin, calretinin, four splicing variants of ErbB4 mRNA (JM-a, JM-b, CYT-1, CYT-2), pan-ErbB4, and MIAT (see Table S2 in the online
data supplement). All primer pairs showed amplification efficiency above 98% in standard curve analyses and specific single products in dissociation curve analyses. The qPCR amplicons for each ErbB4 splicing variant yielded a single band of the predicted size on a gel (Figure S1 in online
data supplement) and were confirmed to span the alternative exons of ErbB4 transcripts (Figure S2 in online
data supplement). The qPCR reactions were carried out in quadruplicate by using an ABI StepOnePlus Real-Time PCR System (Applied Biosystems, Foster City, Calif.). Beta-actin, glyceraldehyde 3-phosphate dehydrogenase, and cyclophilin A were used as reference genes to normalize the expression levels of transcripts, as these three housekeeping genes have stable levels of expression across both subject groups (
29,
30). The delta cycle thresholds (dCTs) were calculated for each sample by using the geometric mean of the three endogenous reference genes as the normalizing factor. The expression level for each transcript was then calculated as the expression ratio value (expression ratio=2
–dCTs).
Microarray Analysis
Microarray analysis of MIAT expression in parvalbumin interneurons positive for Vicia villosa agglutinin (VVA) was performed as previously described (see Supplementary Methods section in online
data supplement) (
31).
Genotype Analysis
DNA samples from the 62 subject pairs were genotyped as part of the CommonMind Consortium (
http://commonmind.org/WP/) (see Supplementary Methods in online
data supplement).
Statistical Analysis
Two analysis of covariance (ANCOVA) models (paired and unpaired) were used to compare transcript levels in dorsolateral prefrontal cortex gray matter, layer 2 or layer 4, in the two subject groups. The paired ANCOVA model included mRNA level as the dependent variable; diagnosis as the main effect; subject pair as a blocking factor; and brain pH, RNA integrity number, postmortem interval, and tissue storage time as covariates. This paired model accounts both for our attempts to balance diagnostic groups for sex and age and for the parallel tissue processing of both subjects in a pair, but it is not a true statistical paired design. Therefore, we also used an unpaired ANCOVA model that included mRNA level as the dependent variable; diagnosis as the main effect; and sex, age, brain pH, RNA integrity number, postmortem interval, and storage time as covariates. Nonsignificant covariates were not included in the final analyses. Repeated measures models were also used to compare the levels of calretinin, parvalbumin, pan-ErbB4, and ErbB4 splicing variant mRNAs in layers 2 and 4 in the two subject groups (see Supplementary Methods in online data supplement).
The paired and unpaired analyses from the ANCOVA and the repeated measures model produced comparable levels of statistical significance for the main effect of diagnosis on all transcript levels, except for JM-b levels in layer 2. The results from the paired ANCOVA analyses are reported in the main text and summarized in Table S3 in the online
data supplement. For the repeated-measures models, only the main effect of layer on parvalbumin and calretinin mRNA levels within subjects is reported in the main text; the remaining outcomes are summarized in the online Table S3. The potential influence of comorbid factors on the layer-specific levels of ErbB4 splicing variants in schizophrenia subjects was assessed as previously described (
23) (see online Supplementary Methods). Pearson’s correlation analyses were performed to assess the layer-specific associations among the ErbB4 splicing variants and parvalbumin or calretinin levels (see online Supplementary Methods). In the microarray analysis, normalized and log
2-transformed signals of MIAT were compared in the two subject groups by using paired t tests, as previously described (
31). In the genotype analyses, analysis of variance models were used to test the main effect of genotype on the JM-a/JM-b variant ratio and the CYT-1/CYT-2 variant ratio in the total gray matter of 62 subject pairs or in layer 4 of 39 subject pairs.
Results
Dysregulation of ErbB4 Splicing in Schizophrenia
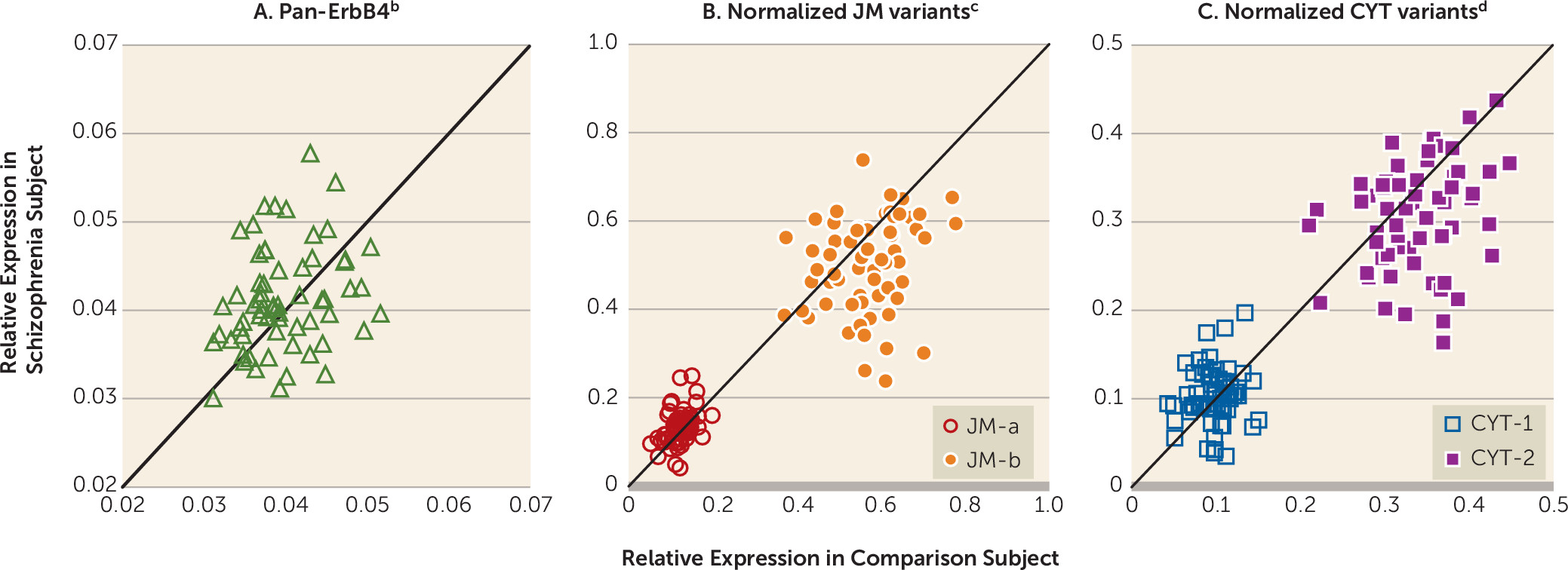
In dorsolateral prefrontal cortex gray matter homogenates from the 62 pairs of schizophrenia and healthy comparison subjects, mean pan-ErbB4 mRNA levels did not differ between subject groups (
Figure 1A). Consequently, we normalized the levels of ErbB4 splicing variants to the pan-ErbB4 levels within each subject to reduce the variability in ErbB4 splicing variant levels attributable to between-subject differences in total ErbB4 expression. The mean normalized JM-a mRNA levels were significantly higher (+10.9%), and the mean normalized JM-b mRNA levels were significantly lower (–11.5%) in the schizophrenia subjects than in the healthy comparison subjects (
Figure 1B). Although the mean normalized CYT-1 mRNA levels did not differ between subject groups, the mean normalized CYT-2 mRNA levels were significantly lower (–10.0%) in the schizophrenia subjects (
Figure 1C). None of the covariates was significant in any of these analyses except that RNA integrity number had an effect on CYT-2 mRNA levels (F=5.02, df=1, 60, p=0.03).
Calretinin and Parvalbumin Interneuron Enrichment of Layers 2 and 4, Respectively
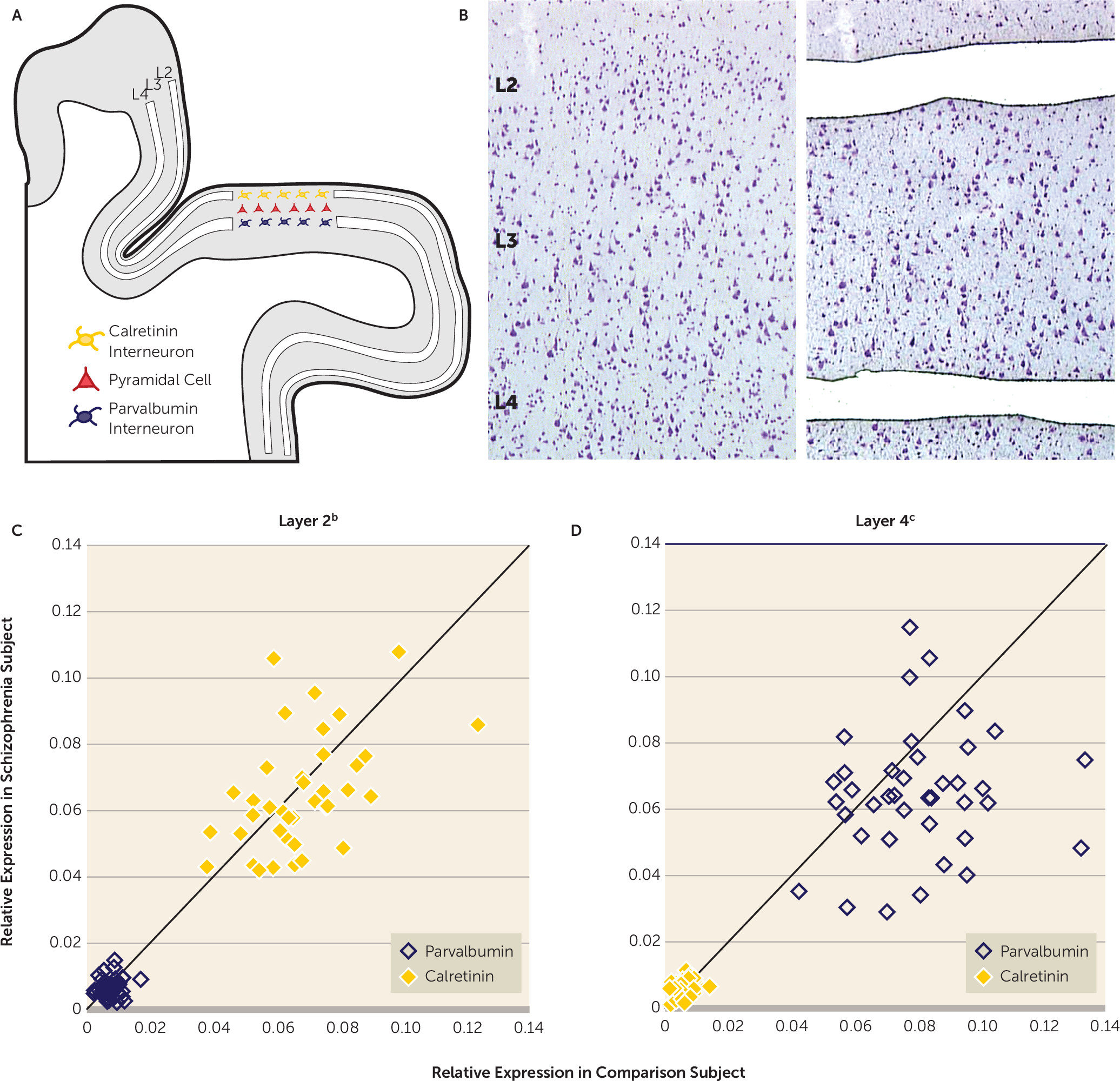
Next we quantified calretinin and parvalbumin mRNA levels in microdissected samples of cortical layers 2 and 4 from the 39 matched pairs of schizophrenia and comparison subjects (
Figures 2A and
2B). Relative to layer 4, calretinin mRNA levels in layer 2 were significantly higher (9.4- and 9.8-fold, respectively) in the comparison subjects (repeated measures: F=593, df=1, 114, p<0.001) and the schizophrenia group (repeated measures: F=517, df=1, 114, p<0.001). Relative to layer 2, parvalbumin mRNA levels in layer 4 were significantly higher (13.9- and 10.9-fold, respectively) in the comparison group (repeated measures: F=590, df=1, 114, p<0.001) and the schizophrenia group (repeated measures: F=373, df=1, 114, p<0.001). These data confirm that layer 2 is largely devoid of parvalbumin interneurons and heavily enriched in calretinin interneurons, whereas the opposite is true for layer 4. Mean calretinin mRNA levels did not differ between groups in either layer 2 or 4 (
Figures 2C and
2D). In contrast, mean parvalbumin mRNA levels were significantly lower (–19.5%) in layer 4 of the schizophrenia subjects but did not differ between groups in layer 2 (
Figures 2C and
2D). In concert, these findings are consistent with a prior report that in the dorsolateral prefrontal cortex of schizophrenia subjects parvalbumin expression is selectively altered in layer 4 but not in layer 2, whereas calretinin expression is not altered in any layer (
20).
Selective Shifts in ErbB4 Splicing From Major to Minor Variants in Layer 4 in Schizophrenia
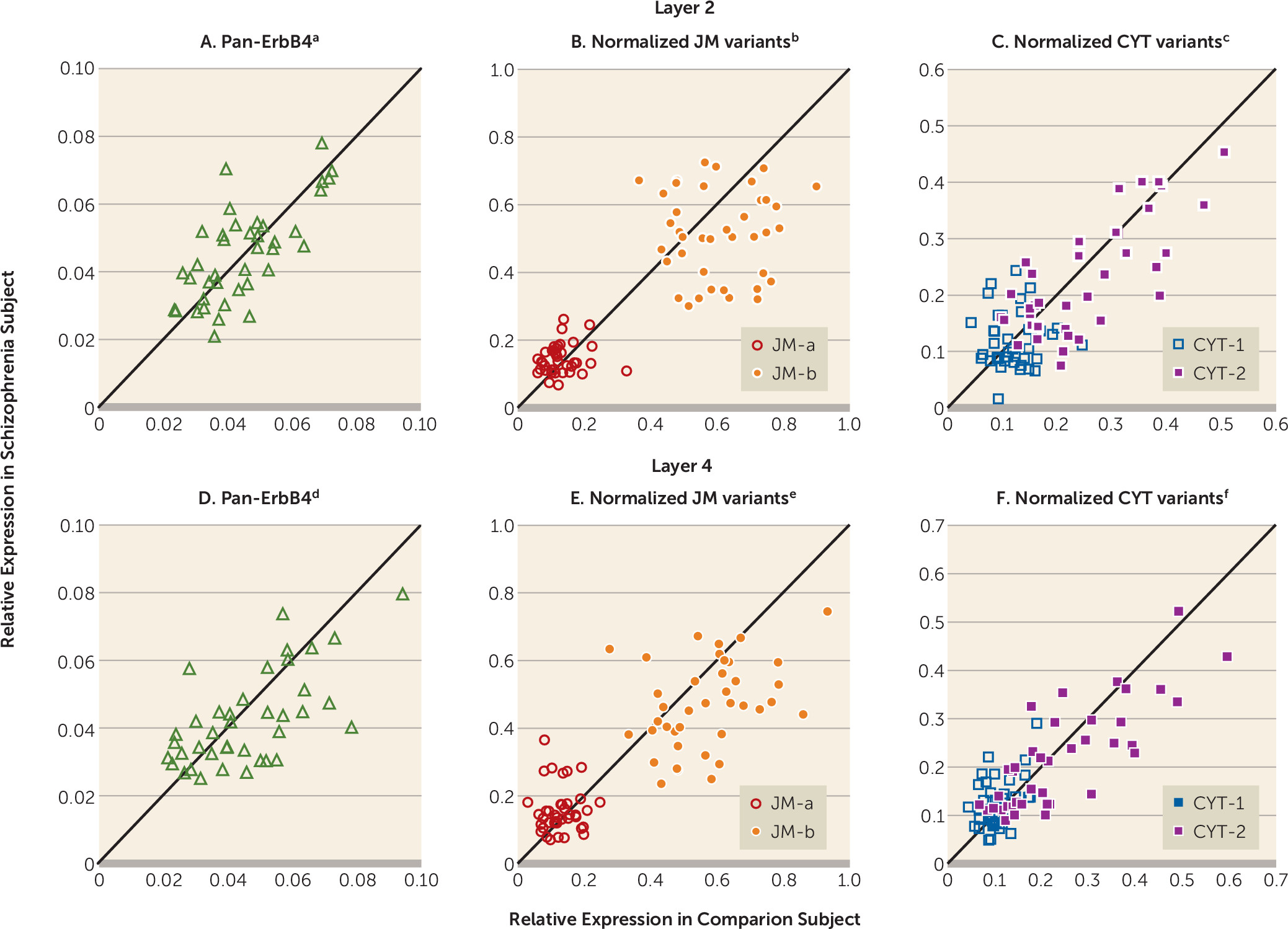
Mean pan-ErbB4 mRNA levels did not differ between subject groups in either layer 2 or layer 4 (
Figures 3A and
3D). In layer 4, mean normalized JM-a mRNA levels were significantly higher (+22.1%), whereas mean normalized JM-b mRNA levels were significantly lower (–17.0%) in the schizophrenia subjects (
Figure 3E). Neither transcript level was significantly different between subject groups in layer 2 (
Figure 3B). Mean normalized CYT-1 mRNA levels were significantly higher (+19.3%), whereas mean normalized CYT-2 mRNA levels were nearly significantly lower (–10.2%) in layer 4 of the schizophrenia subjects (
Figure 3F). Neither transcript level was significantly different between subject groups in layer 2 (
Figure 3C). None of the covariates was significant in any of these analyses except that brain pH had an effect on JM-b mRNA levels in layer 2 (F=8.34, df=1, 37, p=0.006). In addition, mean normalized levels of JM-a, JM-b, CYT-1, and CYT-2 mRNAs in layer 4 of the schizophrenia subjects did not differ significantly as a function of sex, diagnosis of schizoaffective disorder, history of substance dependence or abuse, nicotine use at the time of death, use of antipsychotics, antidepressants, or benzodiazepines, and/or sodium valproate at the time of death, or death by suicide (see Figure S3 in the online
data supplement).
Negative Correlation Between Parvalbumin mRNA Levels and JM-a/JM-b Variant Ratio in Layer 4 in Schizophrenia
In layer 2, calretinin mRNA levels did not correlate with either the JM-a/JM-b variant ratio or the CYT-1/CYT-2 ratio in either the schizophrenia or healthy comparison subjects. In layer 4, a strong negative correlation was observed between parvalbumin mRNA levels and the JM-a/JM-b ratio in the schizophrenia subjects only (r=−0.50, N=39, p<0.001). The CYT-1/CYT-2 ratio did not correlate with parvalbumin mRNA levels in layer 4 of either subject group (see online Table S4).
Preferential MIAT Increase in Parvalbumin Interneurons in Schizophrenia
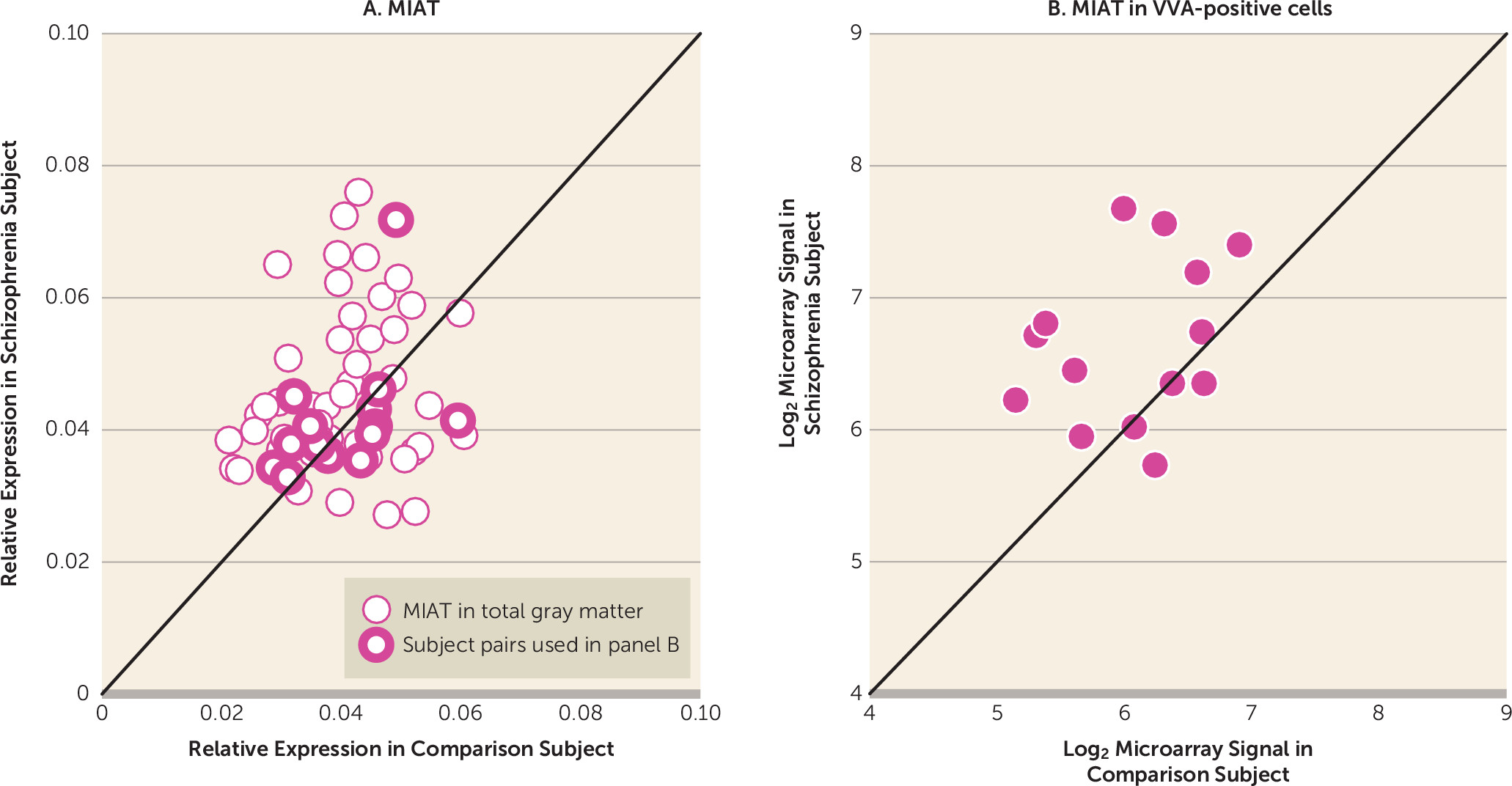
Next we assessed whether abnormal MIAT levels might contribute to dysregulated ErbB4 splicing in schizophrenia. In dorsolateral prefrontal cortex gray matter homogenates from the 62 subject pairs, mean MIAT levels were significantly higher (+12.6%) in the schizophrenia subjects relative to the comparison subjects (
Figure 4A). None of the covariates were significant except for an effect of RNA integrity number (F=4.08, df=1, 60, p<0.05). In dorsolateral prefrontal cortex gray matter, the levels of MIAT were significantly positively correlated with the JM-a/JM-b ratio (r=0.26, N=124, p=0.004) and the CYT-1/CYT-2 ratio (r=0.21, N=124, p=0.03), and they were significantly negatively correlated with parvalbumin mRNA levels (r=−0.20, N=124, p=0.03) across all subjects. MIAT not only is expressed in layers 2 and 4 but also is found in high levels in layers 3 and 5 (
32), suggesting that MIAT may be expressed in both pyramidal cells and interneurons in dorsolateral prefrontal cortex layer 4. Thus, in order to assess MIAT levels selectively in parvalbumin interneurons, we evaluated MIAT levels in laser-microdissected cells labeled with the parvalbumin-specific marker Vicia villosa agglutinin (VVA) from available microarray data in 14 of our subject pairs (online Table S1) (
31). Mean MIAT levels were significantly higher (+50.8%) in VVA-positive cells in subjects with schizophrenia than in healthy comparison subjects (
Figure 4B). In the dorsolateral prefrontal cortex gray matter of these 14 subject pairs, mean MIAT levels did not differ between groups (F=0.17, df=1, 13, p=0.69) (
Figure 4A), suggesting that increased MIAT expression in schizophrenia preferentially occurs in parvalbumin neurons.
Lack of Association Between Schizophrenia-Associated Intronic SNPs at ErbB4 Locus and Ratios of ErbB4 Splicing Variants
Finally, we investigated the effect of schizophrenia-associated intronic SNPs at the ErbB4 locus on the ratios of ErbB4 splicing variants in the dorsolateral prefrontal cortex total gray matter of 62 subject pairs and in dorsolateral prefrontal cortex layer 4 of 39 subject pairs. The risk SNP at rs4673620 was previously implicated with higher JM-a mRNA levels, whereas the risk SNPs at rs7598440, rs707284, and rs839523 were associated with higher CYT-1 mRNA levels in schizophrenia subjects (
4). Our SNP analyses did not reveal a main effect of rs4673620 on the JM-a/JM-b variant ratio in total gray matter or in layer 4 of all subjects. No main effect of rs7598440, rs707284, or rs839523 on the CYT-1/CYT-2 ratio in either total gray matter or in layer 4 was observed in all subjects. Interactions between diagnosis and genotypes were also not detected in either total gray matter or layer 4 of all subjects (see Table S5 in online
data supplement).
Discussion
Dysregulated alternative splicing of ErbB4 mRNAs has been consistently observed in the dorsolateral prefrontal cortex of schizophrenia subjects (
3–
6). Higher levels of the JM-a variant and unaltered pan-ErbB4 levels in schizophrenia subjects have been replicated in all prior studies and were also confirmed in the present study. In contrast to prior studies, we also detected the expected lower levels of JM-b mRNA in schizophrenia subjects. Reciprocal differences in the levels of JM variants, accompanied by unchanged pan-ErbB4 mRNA levels, argue for an abnormal shift in ErbB4 splicing from the JM-b to JM-a variant in schizophrenia. Three previous reports showed higher CYT-1 variant levels in the dorsolateral prefrontal cortex of schizophrenia subjects (
3,
4,
6), whereas the most recent study found no change (
5). We found unaltered CYT-1 variant levels but did detect lower levels of CYT-2 variants in schizophrenia subjects. In concert, these findings suggest that a modest shift in splicing from CYT-2 to CYT-1 variants may occur in some subjects with schizophrenia.
Next we sought to investigate whether dysregulated ErbB4 splicing is associated selectively with parvalbumin interneurons and contributes to lower parvalbumin mRNA levels in schizophrenia. The laminar microdissection yielded preferential enrichment of calretinin and parvalbumin mRNA in layer 2 and layer 4, respectively, and comparable levels of pan-ErbB4 in both layers. Because ErbB4 is localized in calretinin and parvalbumin interneurons in the primate dorsolateral prefrontal cortex (
26), our results support the idea that the layer-specific quantification of ErbB4 mRNAs allows an indirect measurement of their levels in each of those cell types. Pan-ErbB4 mRNA levels were unaltered in either layer 2 or 4 of the schizophrenia subjects, suggesting that transcription of the ErbB4 gene is not affected in either calretinin or parvalbumin interneurons in schizophrenia. We found significantly higher levels of JM-a and CYT-1 variants and significantly lower levels of JM-b variants and nearly significantly lower CYT-2 variant levels in layer 4, but not in layer 2, of the schizophrenia subjects. These findings suggest that the dysregulated shift in ErbB4 splicing from major to minor variants occurs selectively in parvalbumin interneurons. Finally, the negative correlation between the JM-a/JM-b variant ratio and parvalbumin mRNA levels selectively in layer 4 of the schizophrenia subjects supports the hypothesis that dysregulated splicing at the JM locus contributes to an activity-dependent down-regulation of parvalbumin expression in schizophrenia.
In layer 2 we detected nonsignificantly higher JM-a and lower JM-b levels in schizophrenia subjects, indicating a weak shift in splicing from JM-b to JM-a variants. Interestingly, ErbB4 is also present in most of the cholecystokinin-positive (CCK) interneurons as well as in approximately 10% to 30% of calbindin-positive interneurons, which together comprise about 18% of ErbB4-positive cells in the superficial layers (but only approximately 5% in the middle layers) of primate dorsolateral prefrontal cortex (
26). The majority of calbindin-positive cells also express somatostatin (
33). Therefore, although the majority of ErbB4 transcripts quantified in layer 2 are from calretinin interneurons, some proportion of these ErbB4 transcripts would likely be from CCK and somatostatin interneurons. Interestingly, both CCK and somatostatin mRNA levels are lower in the dorsolateral prefrontal cortex of schizophrenia subjects (
34), suggesting that the modest shift in splicing at the JM locus in layer 2 of schizophrenia subjects may represent dysregulated ErbB4 splicing in these cell populations.
The noncoding RNA MIAT was recently reported to regulate the alternative splicing of ErbB4 transcripts (
7). In dorsolateral prefrontal cortex total gray matter homogenates of our 62 subject pairs, MIAT levels were higher in schizophrenia subjects, positively correlated with the ratios of minor to major variants, and negatively correlated with parvalbumin mRNA levels across all subjects. These findings implicate higher MIAT levels as a possible mechanism responsible for the abnormal shifts in ErbB4 splicing in parvalbumin interneurons in schizophrenia. In support of this idea, MIAT levels were higher in VVA-positive parvalbumin interneurons but unaltered in total gray matter homogenates of the schizophrenia subjects in the 14 subject pairs used for both the microarray and the qPCR analyses, demonstrating that MIAT levels are higher preferentially in parvalbumin interneurons in schizophrenia.
Converging evidence suggests that ErbB4 signaling positively regulates the activity of parvalbumin interneurons. ErbB4 signaling increases the formation of excitatory inputs onto parvalbumin interneurons by regulating the synaptic accumulation of postsynaptic density protein 95 (PSD-95) (
15,
16) and the AMPA receptor subunit GluA1 (
35). Moreover, ErbB4 activation inhibits the activity of the Kv 1.1 voltage-gated potassium channel in parvalbumin interneurons (
36) and decreases postsynaptic inhibitory currents by internalizing GABA
A α1 receptors (
37). Previous literature suggested that higher JM-a levels might lead to the loss of ErbB4 signaling, given the differential downstream consequences of the JM isoforms (
3–
5). Our data suggest that the parvalbumin-specific dysregulated splicing at the JM locus produces a gain of the JM-a isoform as well as a loss of the JM-b isoform in schizophrenia, which may additively contribute to impaired ErbB4 signaling in parvalbumin interneurons.
Our study presents novel evidence for alterations in a molecular cascade that may contribute to parvalbumin interneuron dysfunction in schizophrenia. Specifically, higher MIAT levels in parvalbumin interneurons may lead to an abnormal shift in ErbB4 splicing from the JM-b to the JM-a variant, which would result in impaired ErbB4 signaling. Given the role of ErbB4 signaling in the formation of excitatory inputs, the resulting reduction in excitatory drive to parvalbumin neurons would lead to a corresponding activity-dependent down-regulation of parvalbumin expression in schizophrenia. Lower parvalbumin interneuron activity is associated with reduced cortical network activity at gamma-band frequency (
38), which is required for normal cognitive functions, such as working memory (
39). Indeed, patients with schizophrenia demonstrate lower gamma-band power during working memory tasks (
40). Thus, our findings suggest that dysregulated ErbB4 splicing may provide, at least in part, a molecular substrate for lower parvalbumin interneuron activity that could contribute to impaired gamma oscillations and cognitive dysfunction in individuals with schizophrenia.
Acknowledgments
The authors thank Kelly Rogers and Mary Brady for technical assistance and Drs. Bernie Devlin and Lambertus Klei for providing the genotype information.