Family, twin, and adoption studies have indicated substantial heritability in the etiology of alcoholism, but the mechanisms of transmission are still poorly understood (
1). Genetic animal models have suggested that components of the alcohol dependence syndrome can be genetically and behaviorally separated (
2). It is assumed that interindividual differences in neurotransmitter functions mediating rewarding, aversive, or neuroadaptive effects of drugs underlie individual susceptibility to addiction. Neurobiological studies emphasizing the important role of mesolimbic dopamine systems for reward and reinforcement (
3,
4) have stimulated neurogenetic research with addicted subjects generally focusing on the dopamine D
2 receptor gene (
5).
However, the thesis, based on accumulated data, that mutations of this single gene contribute significantly to the addiction liability of individuals (
6,
7) is still controversial. Thus, variants of other genes involved in central dopamine functions are also under investigation. The dopamine transporter gene (DAT1; locus symbol: SLC6A3) was studied in several groups of addicts (
8–
11) because of the role of the transporter protein in terminating dopaminergic activity in synaptic transmission (
12). Cocaine addicts were of particular interest, as the reuptake protein is known to be a direct target for cocaine (
13). An association of polymorphisms and cocaine dependence could not be demonstrated, but a predisposition for paranoia symptoms was found in a particular subgroup (
8,
9). In alcoholics, the striatal density was shown to be abnormal in studies using single photon emission computed tomography; the observation of opposite abnormalities of receptor densities in violent and nonviolent subjects (
11) may imply that differences in the expression of the genotype account for the phenotypic variation in personality traits of the alcoholics studied.
We recently found a significantly higher prevalence of the nine-repeat (A
9) allele of a variable-number tandem repeat in the 3′ untranslated region of the DAT1 gene in alcoholics who reported histories of withdrawal seizures or delirium than in comparison subjects (
14). The aim of this study was to determine whether this variant is also related to symptoms of uncomplicated alcohol withdrawal (
15).
METHOD
The study was performed at the Department of Psychiatry of the Free University of Berlin, where patients are offered outpatient consultations, subsequent hospital detoxification treatment, and an outpatient rehabilitation program. During the first consultation visit, each subject included in the study gave written informed consent relating to documentation of data on personal history and course of alcohol withdrawal for scientific purposes. After the withdrawal period was completely terminated, additional informed consent was obtained for the subsequent blood sampling for genotyping. The study patients had to meet the criteria for ICD-10 alcohol dependence syndrome after they had been evaluated for demographic and psychiatric history data, including information obtained by using the substance abuse section of the Composite International Diagnostic Interview (
16). In accord with Cloninger's neurobiological learning model, personality traits were assessed by means of the Tridimensional Personality Questionnaire (
17). The study was approved by the Ethics Committee of the Rudolf Virchow University Hospital.
On the day of admission to the inpatient unit each patient underwent psychiatric, neurological, and medical examinations, including biochemical tests for acute and chronic intoxication (blood concentrations of ethanol and carbohydrate-deficient transferrin [18]) and liver function (e.g., γ-glutamyltransferase). We evaluated consecutively admitted patients but restricted our study to men owing to our experience that male alcoholics usually have a higher alcohol intake than do women. Furthermore, for complete assessment of withdrawal symptoms the design included only patients who reported having had their last drink on the day of admission to the hospital and had measurable ethanol blood concentrations. Patients with major psychiatric or somatic disorders and those with histories of illegal drug use were excluded.
Withdrawal symptoms were recorded with the revised Clinical Institute Withdrawal Assessment for Alcohol Scale (
19), a 10-item clinical rating scale focusing on subjective and objective symptoms of withdrawal (each item is rated on a scale ranging from 0 to 7 or 0 to 4). The scale was administered by a nurse every 2 hours. If the score was higher than 12, the patient received 500 mg of clomethiazole orally to treat or prevent dramatic withdrawal symptoms. Symptom check and administration of medication were repeated in a 2-hour scheme until the scores were consistently below 12. For the assessment of the duration of withdrawal, the number of days with clear withdrawal symptoms (i.e., score above 6 on the withdrawal scale) was recorded. Severity of withdrawal was characterized by the total score, defined as the highest per-day scores summed for all days with clear withdrawal symptoms. Intensity of withdrawal was indicated by the peak score, i.e., the highest score that had ever been achieved on a withdrawal day. Amount of withdrawal medication was defined as the accumulated dose of clomethiazole (in milligrams) administered to a patient.
Genotyping
After venous blood samples were drawn, genomic DNA was extracted from anticoagulated or lymphoblastoid cell lines by using a salting-out method (
20). Allelic variants were characterized by determining a 40-base-pair variable-number tandem repeat in the 3′ untranslated region on chromosome 5p15.3 (
21). DNA amplification by polymerase chain reaction of the 40-base-pair repeat was performed as described elsewhere (
22). The polymerase chain reaction products were separated by 10% polyacrylamid gel electrophoresis and were then silver stained, and fragment sizes were determined by comparison with molecular weight standards. The numeric designation of each allele referred to the number of repeats (A
n) it contained. Genotyping was carried out without knowledge of the clinical status of the patients.
Subjects
A total of 48 patients were included in the study. Twenty-six of subjects were typed A
10/A
10, and 22 were characterized A
9/A
10. All subjects were men aged 22–58 years and had primary alcoholism (
23).
Statistical Analysis
Differences in patient variables (i.e., history data and single symptom scores on the Clinical Institute Withdrawal Assessment for Alcohol Scale) between the two subgroups were tested by means of chi-square statistics and Student's t test by SPSS (
24). For full utilization of the withdrawal data and avoidance of multiple testing, variables indexing withdrawal, i.e., withdrawal severity, intensity, duration, and medication, were factor analyzed (principal component analysis and varimax rotation) by SPSS (
24). Composite factor scores based on the weights assigned by factor analysis to each of the four clinical variables indexing withdrawal were then subsequently tested for significant differences by means of Student's t test by SPSS (
24). For further assessment of the relevance of the genotype in comparison to that of three clinical variables (i.e., age at onset of alcoholism, expressed as the time of first loss of control over alcohol intake; amount of alcohol per day in the month preceding hospital admission; ethanol blood concentration at hospital admission), withdrawal (defined as the general factor resulting from factor analysis) was examined as the main outcome criterion (and dependent variable) by means of multiple regression analysis (stepwise) by SPSS (
24). Results are presented for the 0.05 level of significance.
RESULTS
The patients in the two genotypically defined subgroups did not differ significantly in any demographic or personal variable (including family history of alcoholism). However, alcoholic delirium or seizures were reported in the patient histories of significantly more of the patients carrying the A
9 allele (50.0%, N=11) than those without the A
9 allele (19.2%, N=5) (χ
2=3.78, df=1, p=0.05 with Yates's correction). The groups did not differ significantly in biochemical indices of acute and chronic intoxication at time of admission for hospital detoxification (
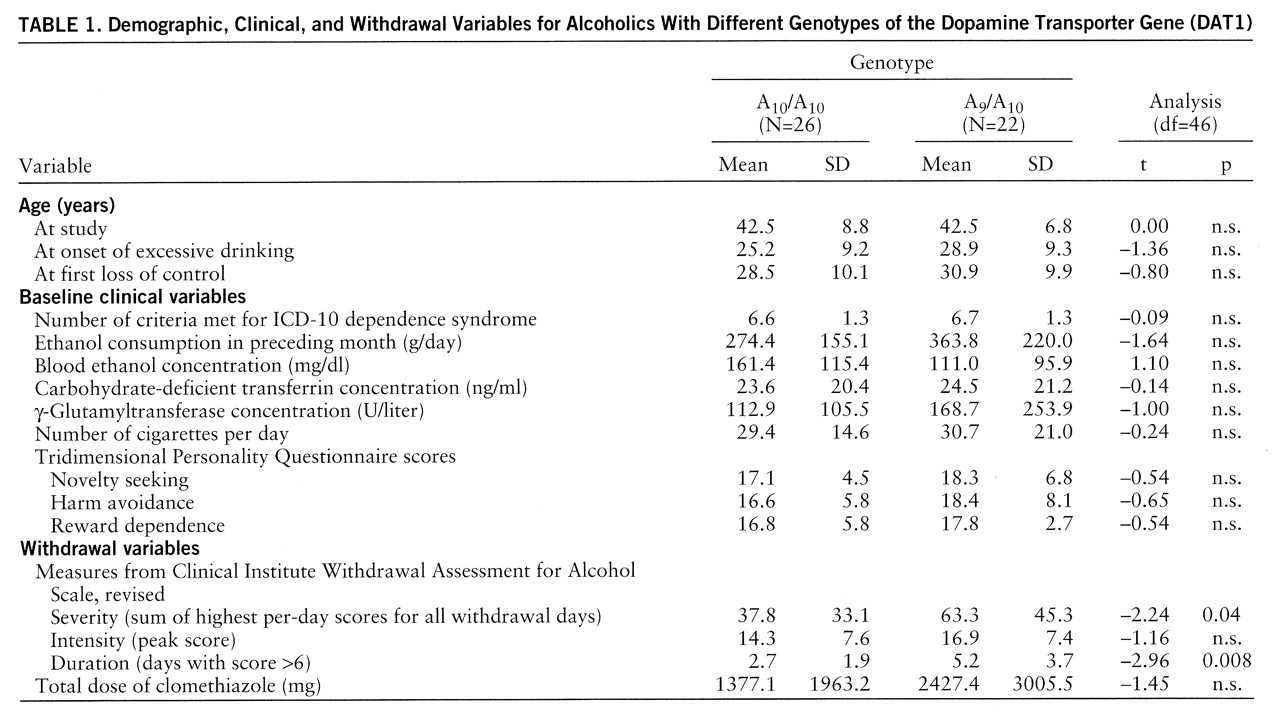
table 1). The values for all variables involving withdrawal symptoms were higher in the group of A
9 carriers, reaching significance for withdrawal severity and duration (
table 1).
Factor analysis of the data indexing withdrawal severity, intensity, duration, and medication resulted in a one-factor solution (general factor “withdrawal”) with an eigenvalue of 3.02 explaining 75.7% of the total variance; subsequent testing of factor scores revealed a significant difference between the patients with the A10/A10 genotype and those carrying the A9/A10 genotype (–0.29 versus 0.34) (t=–2.20, df=46, p=0.03), indicating a more severe withdrawal syndrome in the A9 carriers than in the patients without this variant.
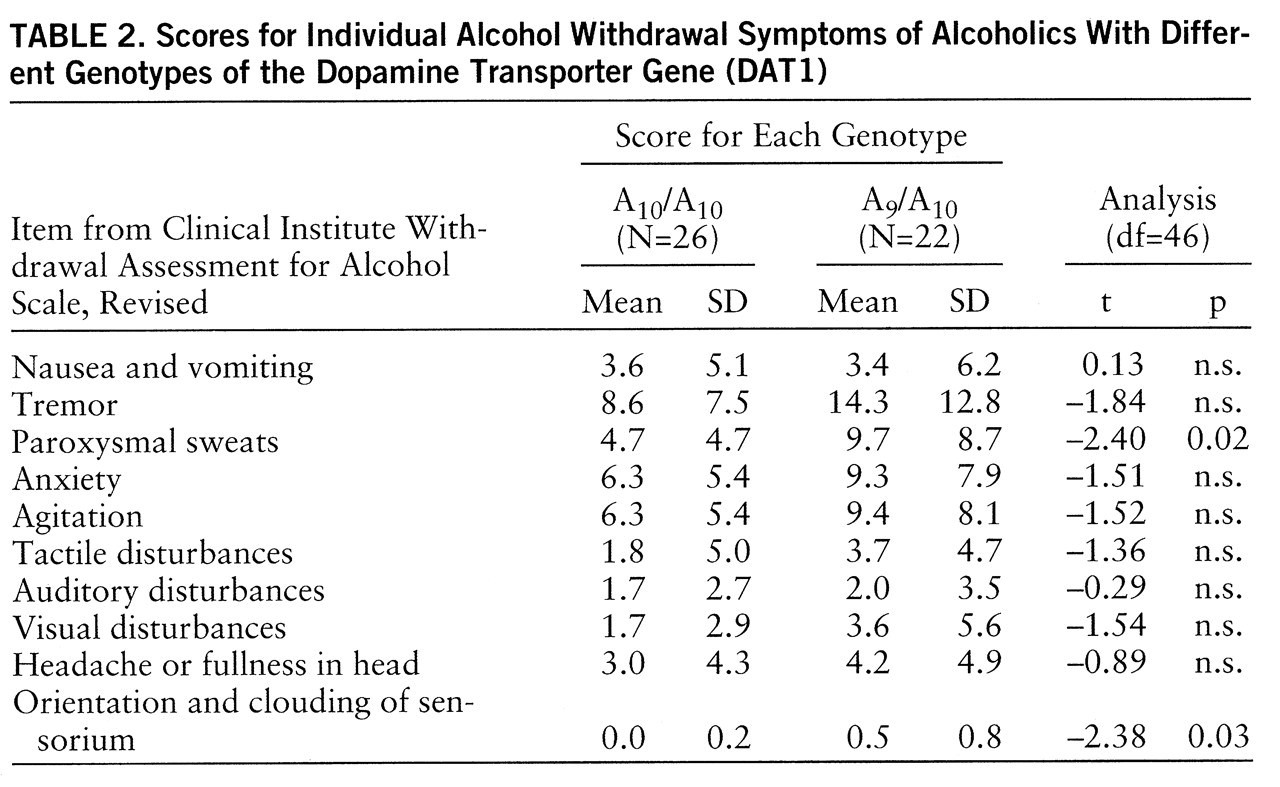
Analysis of single symptoms in the revised Clinical Institute Withdrawal Assessment for Alcohol Scale also revealed higher scores on all symptoms for the A
9 carriers than for the comparative group; significant differences, however, were obtained only for the symptoms “paroxysmal sweats” and “orientation and clouding of sensorium” (
table 2).
Multiple regression analysis performed to assess the relevance of the genotype in comparison to that of major clinical variables yielded a significant model (adjusted R2=0.20, F=8.63, df=1,39, p=0.005). The stepwise procedure resulted in beta weights only for amount of alcohol (β=0.42, p=0.005) and DAT1 genotype (β=0.24, p=0.10), thus indicating that about 20% of the variance in withdrawal symptoms could be explained by drinking style (16%) and the allelic variant (4%) studied in our patients.
DISCUSSION
The main finding of the study is that different withdrawal syndromes emerged in two genetically determined subgroups of alcoholics. The withdrawal syndromes were more pronounced in subjects carrying the nine-copy variable-number tandem repeat of the DAT1 gene than in individuals without this variant. After the variables predicting withdrawal were ranked, however, the amount of ethanol consumption was the most informative data source, as indicated by the significance of ethanol consumption in our model; this finding also fits well with general clinical experience (
25). The genotype was revealed to be less relevant; the statistical evaluation pointed to only marginal significance for the individual effect of DAT1.
In the design of the study, monitoring the patients' clinical picture prospectively and assessing the genotype independently were essential for ensuring data validity. Both groups were homogeneous for clinical variables such as sex, age at study and age at onset of alcoholism, drinking pattern, degree of acute and chronic intoxication, and a defined and parallel start of detoxification. The patients had primary, pure alcoholism (
23), and such alcoholics typically have no experience with illegal drugs in Germany. As our patients differed regarding history of withdrawal delirium and seizures and, as now shown, uncomplicated withdrawal symptoms, we conclude that the genetic variant might be involved in common mechanisms underlying alcohol withdrawal, contributing to different phenotypes. We propose that the DAT1 genotype is particularly involved in neuroadaptation, especially in the brain's capacity to compensate for long-term effects of ethanol on cerebral function. These data extend the results of our recent association study (
14), suggesting that alcohol-dependent patients with the A
9 allele of the DAT1 gene are more maladaptive, or vulnerable to alcohol withdrawal symptoms, than are individuals lacking this variant. The relevance of the dopamine transporter system for alcoholism was further demonstrated by a Japanese association study showing a greater frequency of the A
7 allele of the DAT1 gene in alcoholics with an inactive ALDH2 gene (
26).
The functional consequence of the nine-copy DAT1 repeat is not yet known. Although it is unlikely that the nine-repeat motif itself in the 3′ untranslated region of DAT1 has functional relevance, it is close to the coding region and could be in linkage disequilibrium with a vulnerability-causing mutation that alters gene expression or protein structure. Therefore, in analogy to binding studies (
27), it could be speculated that the greater withdrawal symptoms experienced by our patients might reflect hyperdopaminergic states due to insufficient clearance of dopamine in the synaptic cleft (
12) after cessation of chronic ethanol consumption. A small proportion of the variance in withdrawal phenomena (20%) was explained by the DAT1 genotype and an exogenous factor (amount of ethanol consumption). This observation points to a complex gene-environment interaction that permits the assumption that other gene polymorphisms regulating neurotransmitter systems, e.g., dopamine D
2 receptors (
28) or the γ-aminobutyric acid (
29), noradrenaline (
30), or glutamate (
31) system, also contribute to withdrawal vulnerability in a syndromal and continuous manner, as is typical for quantitative trait loci (
2).
Furthermore, it is interesting that the dopamine transporter system may also be involved in the adaptation to certain nondrug (environmental or internal) stimuli corresponding to temperamental or personality variables (such as habitually violent or nonviolent behavior [11]) or to certain cerebral processing disturbances (e.g., attention deficit disorder [32]), conditions that have been found to predispose to alcoholism (
33,
34). No association between the genotype and personality scores on the Tridimensional Personality Questionnaire was found in our patients. However, there is evidence that inherited variants of the DAT1 gene may influence vulnerability to other psychiatric diseases as well (
35). Since the dopamine transporter system is known to be a target of psychotropic drugs, especially cocaine (
13), it might also be involved in cerebral adaptation to these drugs on functional (
10) and experimental (
36) levels. A relationship between the A
9 allele and a clinical phenotype has been shown for cocaine-induced paranoia (
9); the present study demonstrated a similar relationship in alcohol withdrawal.