The etiology of severe eating disorders—anorexia nervosa, bulimia nervosa, and binge eating disorder—is complex, and multiple influences confer risk for this behavior
(1). As noted by Kaye and Strober
(2), the stereotyped clinical presentation, sex distribution, and age at onset support the likelihood that there is some biological vulnerability to this disorder. Large twin studies show that the co-twin of a twin affected with anorexia nervosa is 26 times as likely to have a lifetime diagnosis of bulimia nervosa as is the co-twin of an unaffected twin
(3), strengthening the notion that genes are important in the etiology of anorexia nervosa and bulimia nervosa.
An interesting candidate gene that makes “biological sense” for a role in eating disorders and has not yet been examined, to our knowledge, is the gene for insulin-like growth factor 2. This gene,
IGF2, is located on chromosome 11p15.5
(5). Insulin-like growth factor 2, also known as somatomedin A, is a single-chain polypeptide that shares an amino acid sequence homology of about 47% with insulin and about 31% with relaxin and with them comprise the insulin family of polypeptide growth factors. Their functions include mediation of growth hormone action, stimulation of growth of cultured cells, stimulation of the action of insulin, and involvement in development and growth. They appear to be autocrine regulators of cell proliferation. Consistent with this profile of metabolic actions, an association has been reported between a single nucleotide polymorphism (SNP) in the 3′ untranslated region of the
IGF2 gene (
ApaI) and body mass index
(6) that accounts for a small percentage of the population variance. A number of studies appear to confirm this initial finding, including suggestive evidence from a genome scan for obesity
(7–
10).
IGF2 is also expressed in fetal and adult brains
(11,
12).
The transmission disequilibrium test
(13) and its extension to quantitative traits
(14,
15) provide an efficacious procedure for detecting linkage and association, especially in the presence of population admixture. Even though this procedure has been widely used, it is not suitable for groups composed of families with multiple siblings, which decrease the efficiency of the analysis performed. More recently, a new unified approach, the family-based association test
(16), for testing association by using any type of family configuration and any type of phenotype (either qualitative or quantitative) has been developed. As our subject group was composed of families with multiple siblings, we applied this new procedure, which allowed us to use all the information contained in our study group despite the varied number of siblings in these families. In this large, nonclinical group of 376 nuclear families we used the family-based association test
(16) to test the association between three
IGF2 SNPs and eating behavior. Eating behavior was assessed by using the 26-item Eating Attitudes Test
(17,
18), which is probably the most widely used standardized measure of symptoms and concerns characteristic of eating disorders. Numerous studies have used the Eating Attitudes Test as a screening tool. Moreover, overall scores show heritability of approximately 40% in normal female twins
(19).
Results
The two self-report measures of eating behavior were strongly correlated in these subjects (N=845): the score on the Eating Attitudes Test correlated with both the Eating Disorder Inventory-2 body dissatisfaction subscale (r=0.63, p<0.001) and drive for thinness subscale (r=0.88, p<0.001). Similar results were observed when men and women were separately analyzed. Body mass index was also correlated with Eating Attitudes Test scores, but weakly (r=0.13, p<0.001).
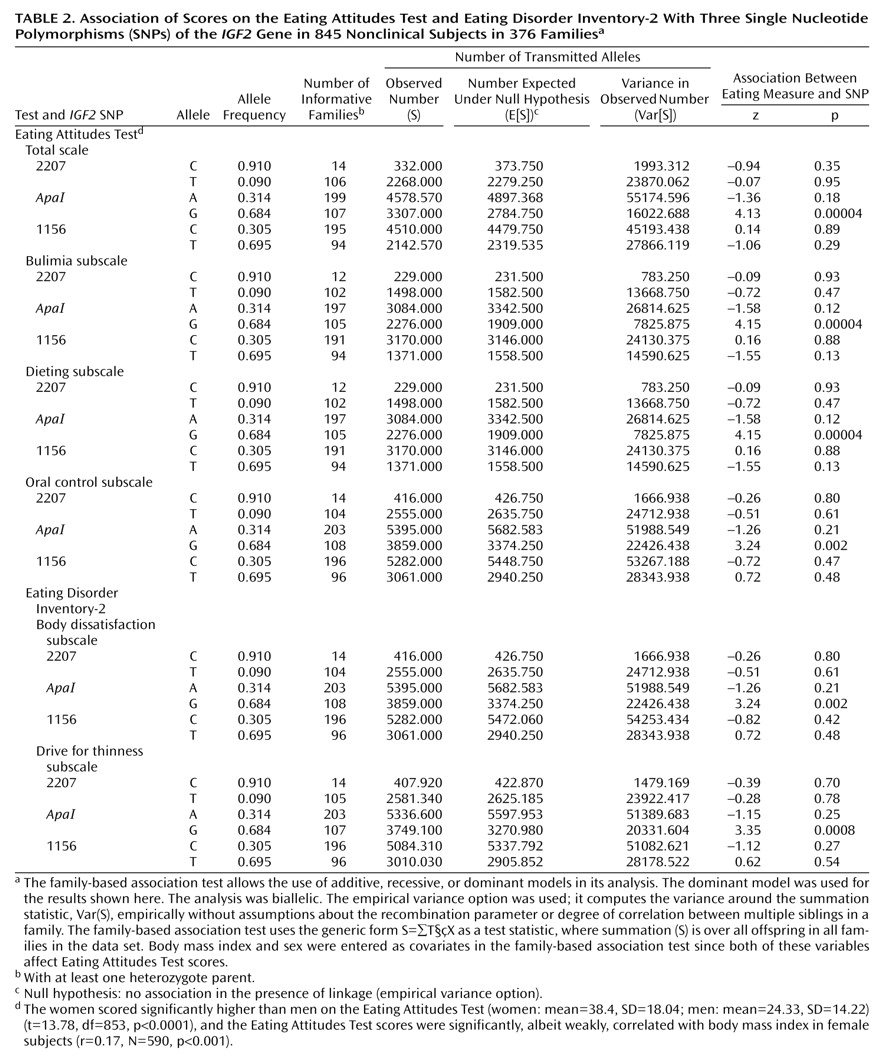
As shown in
Table 2, analysis with the family-based association test showed a highly significant association between the
IGF2 ApaI G allele and Eating Attitudes Test scores overall as well as for each of its subscales: bulimia, dieting, and oral control. Significant association was also observed between the
IGF2 ApaI G allele and the scores on the body dissatisfaction and drive for thinness subscales of the Eating Disorder Inventory-2. Subjects with the G allele had higher scores. Haplotype analysis as provided for in the program for the family-based association test showed that the association with the CAT haplotype (2207-
ApaI-1156) was significant for scores on the total Eating Attitudes Test (z=–3.04, p=0.003) and all three subscales: bulimia (z=3.05, p=0.002), dieting (z=3.05, p=0.002), and oral control (z=–2.01, p=0.05). This haplotype was associated with lower Eating Attitudes Test scores. Note, however, that the genetic information is primarily generated by the
ApaI SNP, whereas little additional information is provided by the two other SNPs genotyped in this study, 1156 and 2207.
The data were also analyzed with a qualitative approach by setting a cutoff point on the Eating Attitudes Test of 20. Individuals scoring higher than 20 on this scale are generally considered at risk for eating disorders
(20), and we categorized subjects with scores above 20 as “affected” in the family-based association test. This also indicated preferential transmission of the
IGF2 ApaI G allele to the group with high scores on the Eating Attitudes Test (z=3.94, p=0.00008). Hence, the quantitative and qualitative approaches to data analysis yielded similar results.
We first estimated the effect size of the
IGF2 ApaI G allele by comparing mean scores of all female subjects with the AA and GG genotypes on the Eating Disorder Inventory-2 drive for thinness subscale (AA: mean=12.71, SD=9.99; GG: mean=16.13, SD=9.09). This difference was significant (t=2.06, df=310, p=0.04, N=312). The effect size of GG homozygosity is small (0.36) according to Cohen’s widely used interpretation
(24). Similar results in regard to effect size were obtained for the other eating-related measures. Small effect sizes have generally been observed for genes contributing to behavioral quantitative trait loci representing a variety of phenotypes. For example, in a meta-analysis of the DRD4 7-repeat allele and attention deficit hyperactivity disorder, an overall odds ratio of 1.9 (corresponding to a small effect size) was observed
(25).
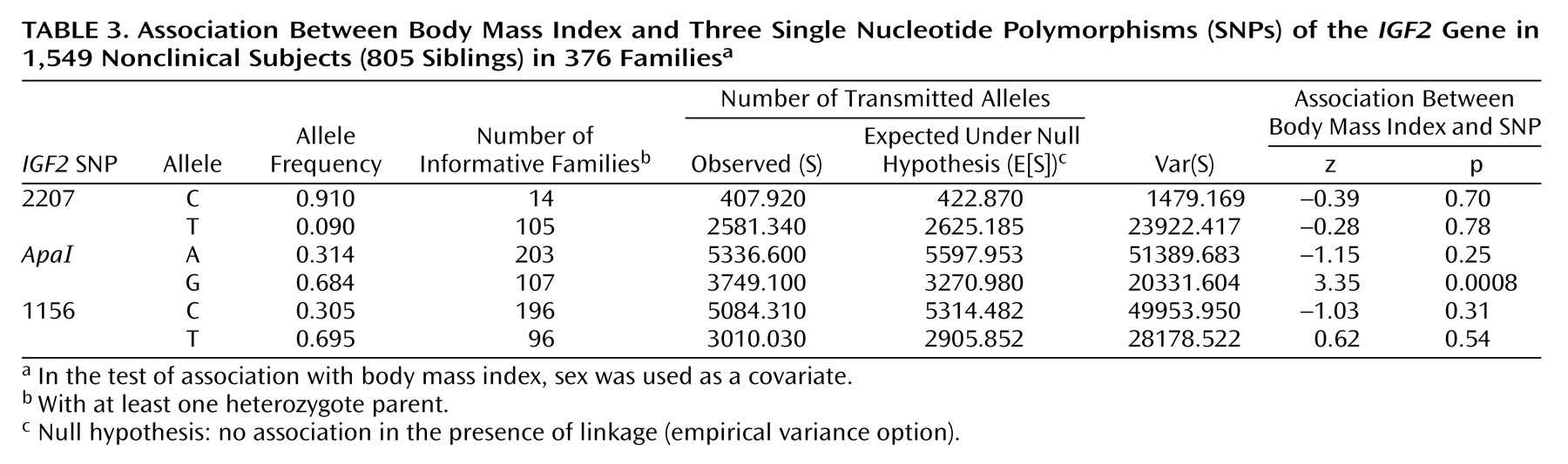
We also tested for association between the three
IGF2 SNPs and body mass index (
Table 3). A highly significant association was observed between the
IGF2 ApaI G allele and body mass index.
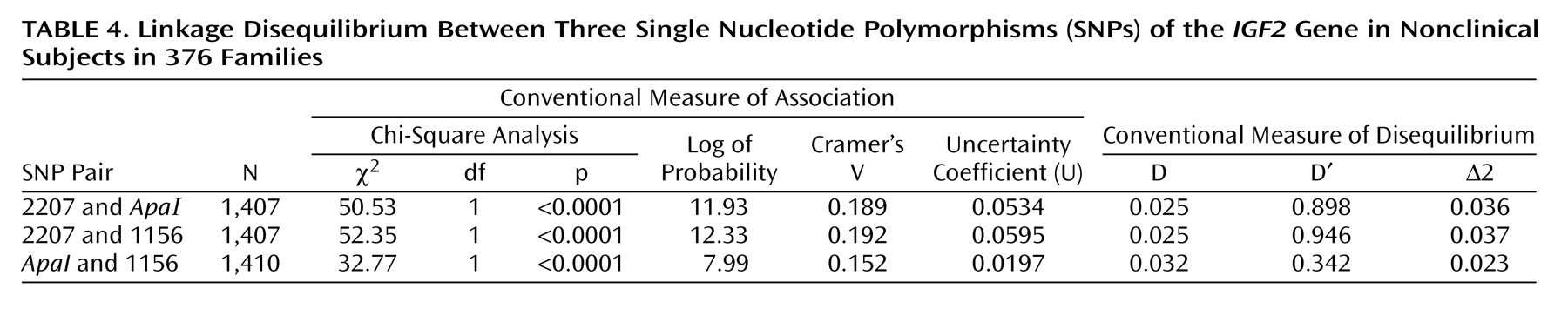
All three SNPs are in linkage disequilibrium (
Table 4).
Discussion
One particular SNP,
ApaI, in the
IGF2 gene has been shown to have a modest influence on body mass index and an inherited predisposition to weight gain
(9). The current finding that it may also contribute to the pathology of eating disorders, as evidenced by its association with scores on the Eating Attitudes Test, is intriguing. Previous studies of this gene in humans have focused on its role in metabolism and body composition. The current findings extend our knowledge of this gene’s action by exploring the impact of the common
IGF2 polymorphisms on psychological processes related to eating behavior. Neurotransmitter modulation of appetitive behavior is the focus of most hypotheses regarding the etiology of severe eating disorders
(26–
28). However, the current results to some measure challenge this view, and we hypothesize that inborn metabolic tendencies to gain weight in some women may trigger constant dieting that in some individuals eventually leads to severe eating disorders, including anorexia nervosa, bulimia nervosa, and binge eating disorder. Not surprisingly, a gene that predisposes to modest increases in body mass index also is also associated with scores on the Eating Disorder Inventory-2 drive for thinness subscale, which measures excessive concern with dieting.
These data confirm in an exclusively Jewish population the association between the
IGF2 ApaI G allele and body mass index that has been observed in several investigations
(6–
10). In a study in the United Kingdom, the less common
ApaI A allele was associated with lower body mass index
(9). In the current study this same allele was also associated with lower body mass index. However, in an American study of a mixed ethnic group of 500 men and women, Caucasian subjects with the
IGF2 ApaI AA genotype exhibited significantly greater fat mass than did Caucasian GG subjects, but no association with body mass index was observed
(8). It is interesting that the evidence provided by the U.K. studies
(5,
6,
9,
10) suggests that the association between
IGF2 SNPs and body mass index represents an association between a genomic region rather than a specific etiological SNP. Moreover, the weight-lowering effect of less common
IGF2 alleles was apparent in all quintiles of body mass index, strengthening the notion that rare
IGF2 SNPs in the U.K. study exert their effects across the whole range of body weights. Overall, these results suggest that the
IGF2 ApaI polymorphism’s role in body composition is influenced by both other SNPs within the
IGF2 genomic region and other genes.
The mechanism by which
IGF2 affects body mass index was suggested in a study employing a 100-day overfeeding protocol conducted with pairs of monozygotic twins
(29). In response to caloric surplus, fasting plasma levels of insulin increased more among the subjects with the
IGF2 ApaI GG genotype than among those with AA or AG. The changes were independent of changes in total fatness. The
ApaI G allele is also associated with significantly higher levels of
IGF2 mRNA than is the A allele, showing a role for this polymorphism in the transcription of this gene
(30). As expected for a hormone that in humans increases growth, the
IGF2 knockout mouse presents a lighter, smaller phenotype
(31) and a transgenic
IGF2 mouse overexpressing insulin-like growth factor 2 is heavier than controls
(32).
In the current study, the
IGF2 ApaI G allele was associated with a number of phenotypes (various measures of eating behavior, body mass index). The association across phenotypes reflecting direct measurements of eating behavior is not surprising since these variables (Eating Attitudes Test, its subscales, Eating Disorder Inventory-2 body dissatisfaction and drive for thinness subscales, body mass index) are correlated. The current results therefore support the notion that the association between the
IGF2 ApaI G allele and various measures of eating behavior and body mass index are “driven” by the G allele’s effect on body mass and composition
(6–
10). It is worth noting that the metabolic profile associated with the
IGF2 ApaI G allele precedes the onset of eating disorders and sets the stage, by its impact on adiposity and body mass index, for the onset of the clinical syndromes associated with eating disorders in some women. Although we have emphasized the risk conferred by the
IGF2 ApaI G allele, it should be underscored that the rare AA homozygote subjects (approximately 9% in this population) are seemingly protected by their low body mass index against the development of eating disorders.
We are aware of at least one other study, of binge eating, that showed a significant association between an eating disorder and a “metabolism” gene, the
MC4R coding region (the leptin binding domain) of the proopiomelanocortin gene (
POMC)
(33). All carriers of
MC4R mutations were given a diagnosis of binge eating disorder, suggesting to the authors that
MC4R is a candidate gene in the control of eating behavior. The current report, based on an investigation of a nonclinical population, suggests that a second “metabolism” gene,
IGF2, also contributes to the control of eating behavior in humans.
Anorexia nervosa, bulimia nervosa, and binge eating disorder present a complex phenotype associated with aberrant eating behaviors, body image distortions, impulse and mood disturbances, hormonal and mineral imbalances, and characteristic temperament and personality traits. Employing a strategy of decomposing the phenotype of severe eating disorders into quantitative trait loci, we show in the present study that the
IGF2 ApaI SNP is associated with scores on the Eating Attitudes Test, an instrument sensitive to pathological eating behavior, in a nonclinical population. Our results suggest the hypothesis that some individuals predisposed to modest increases in body mass index due to common polymorphisms in the
IGF2 gene, and perhaps unduly influenced by the media message that thin is better
(34,
35), embark on a course of constant dieting that slowly evolves into a pattern of abnormal eating behavior and, in a small percentage of such individuals, the full-blown clinical syndromes.