The ability to successfully process and label the emotions of others is crucial to human social interaction (
1). Impaired emotion recognition is a hallmark of a number of psychiatric disorders, including schizophrenia, bipolar disorder, and major depressive disorder (
2,
3). Emotion recognition deficits also occur in neurodegenerative illnesses like Parkinson’s disease and Alzheimer’s disease (
4,
5). Furthermore, there is considerable evidence for individual differences in emotion recognition in healthy populations (
6), as well as evidence that a substantial portion of that variation appears to be under genetic influence (
7). While multiple neural systems putatively subserve emotion recognition, the amygdala appears to have a preferential role in affect processing in both healthy and mentally ill individuals (
1,
8–
10). As the structure of the amygdala is influenced by genetic factors (
11,
12), it is possible that the same genes that influence emotion recognition also influence amygdala volume and vice versa. However, it remains unclear whether the association between amygdala volume and emotion recognition is due to common genetic influences, and if so, which genes in particular are involved. Identifying genes with pleiotropic influence on both traits might reveal those molecular mechanisms that alter brain architecture or function, which in turn affect emotion recognition performance. In an effort to further our understanding of the molecular underpinnings of emotion recognition, our aim in this study was to isolate genes that jointly influence emotion recognition and amygdala volume in randomly selected extended pedigrees.
Numerous neural systems are implicated in emotion recognition, and those systems reside in particular in the frontal and temporal lobes, which together make up the neural pathways responsible for the interpretation of visual emotional stimuli (
9,
10). After a visual stimulus is processed via the lateral geniculate nucleus and the primary visual cortex (V1), the amygdala becomes the focus of further processing in the brain, receiving input from both cortical and subcortical streams (
13). The amygdala was first implicated in emotional capacity by Brown and Shafer (
14), who noted that monkeys with bilateral temporal lobe lesions were rendered tame and docile. Later this type of lesion was linked to emotion processing and, more specifically, to fearful responses (
15,
16). Subsequent work in primates and rodents narrowed the region of interest for emotion processing down to the amygdala (
17,
18). The role of the amygdala in emotion recognition, and in particular the recognition of negative emotions, was confirmed in humans by lesion studies (
19,
20) as well as functional imaging studies (
10,
21,
22). While there is evidence that variation in the serotonin transporter gene influences the amygdala’s response to emotive faces (
23), the complete genetic architecture of amygdala function and structure is largely unknown (
24).
We report here on bivariate linkage and association analysis in a sample of Mexican American individuals from extended pedigrees. Using bivariate linkage analysis, we identified a region of chromosome 4 as being truly pleiotropic for bilateral amygdala volume and emotion recognition performance. Using association analysis of common variants within that linkage region, we identified a gene, PDE5A, as being influential on both traits.
Results
Heritability and Linkage Analysis
Both amygdala volume (h2=0.72, SE=0.07, p=4.95×10−5; mean=3066.72, SD=434.47) and emotion recognition (h2=0.32, SE=0.06, p=5.12×10−10; happy: mean=7.78, SD=1.47; sad: mean=6.15, SD=1.40; fear: mean=6.61, SD=1.36; anger: mean=5.56, SD=2.21; neutral: mean=5.58, SD=4.37) were highly heritable, and a bivariate model indicated significant genetic overlap between the two traits (rhog=0.25, SE=0.13, p=0.048). For amygdala volume and emotion recognition, age and sex were significant covariates (see Table S1 in the data supplement that accompanies the online edition of this article), and hence the effect of age and sex was covaried in all subsequent analyses. The factor score derived for emotion recognition and amygdala volume was also highly heritable (h2=0.45, SE=0.06, p=5.68×10−22), and the factor loadings for both traits were deemed to be significant at the p<0.001 level; goodness-of-fit statistics were not available because of saturation of the model.
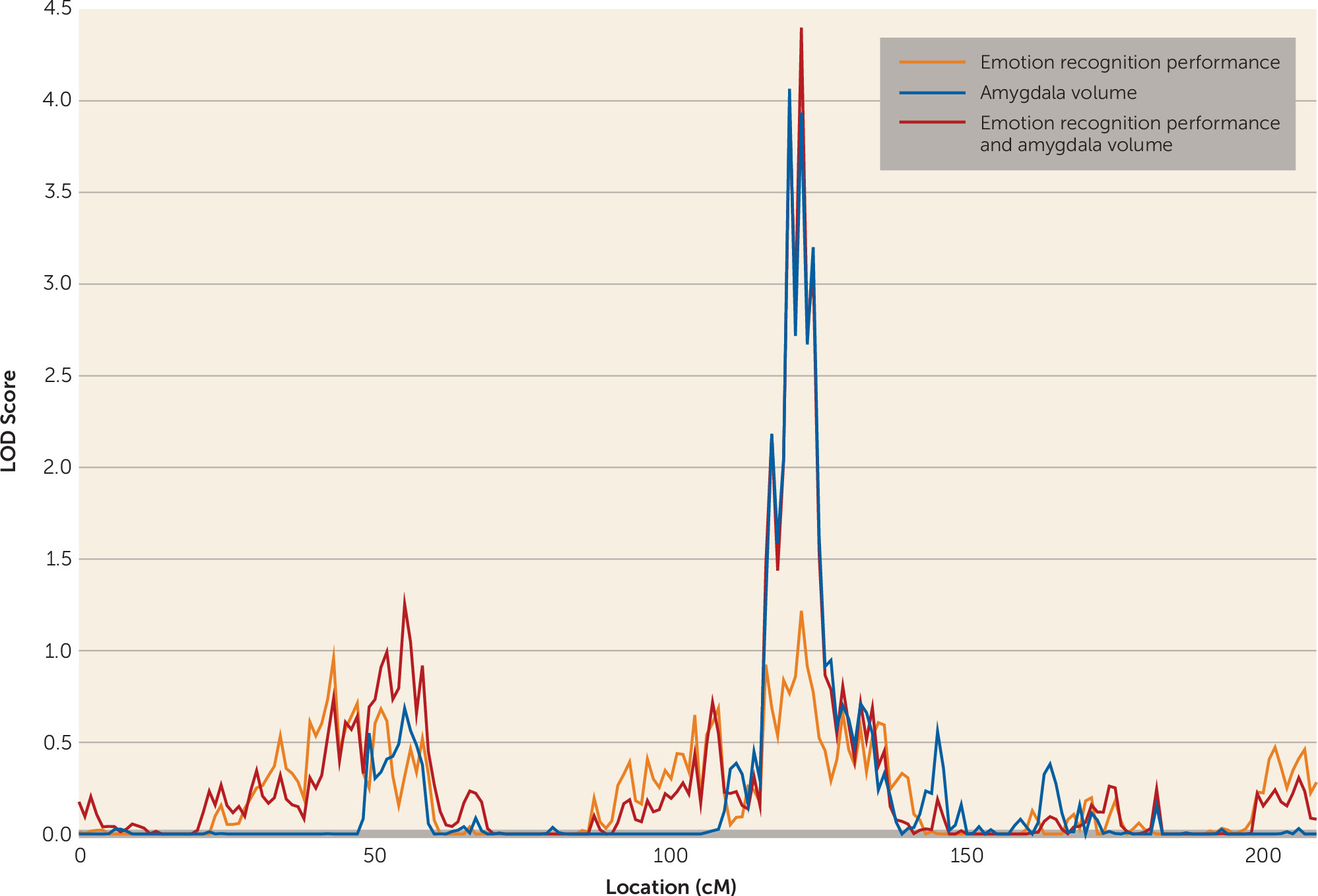
For amygdala volume, one genome-wide significant locus was observed on chromosome 4 at 120 cM (LOD score=4.065). A LOD score of 2.175 was observed for the emotion recognition task on chromosome 2 at 71 cM, meeting criteria for suggestive significance, while the chromosome 4 locus showed some evidence for linkage to emotion recognition (LOD score=1.217) (
Figure 2). Bivariate linkage revealed a genome-wide significant QTL for both amygdala and emotion recognition on chromosome 4 at 122 cM (1 df-equivalent LOD score=4.399), which suggests that this region of chromosome 4 mediates both amygdala volume and performance on the emotion recognition task. The test for pleiotropy versus coincident linkage confirmed the presence of pleiotropy for the two traits at this locus (χ
2=20.12, df=1, p=3.6×10
−6). The factor score, derived from the one-factor model of emotion recognition and amygdala volume, showed genome-wide significant linkage at precisely the same region on chromosome 4 at 122 cM (LOD score=3.336).
The possible confounding role of intracranial volume at this locus was investigated using trivariate linkage. Univariate linkage revealed a QTL of suggestive significance for intracranial volume on chromosome 16 at 37 cM (LOD score=2.65) with little evidence for genetic influence on this trait on chromosome 4 at 122 cM (LOD score=0.47). Moreover, in a trivariate linkage model of the emotion recognition task, amygdala volume and intracranial volume on chromosome 4 at 122 cM showed genome-wide significance (1 df-equivalent LOD score=3.546), and even within this trivariate model, the pleiotropy test supported complete pleiotropy between amygdala and emotion recognition (χ2=7.74, p=2.7×10−3). Furthermore, the possibility that the right or left amygdala might be driving the result was addressed by running univariate and bivariate linkage in the same region. Left amygdala volume (h2=0.7019, SE=0.0788, p=2.03×10−21) had a univariate LOD score of 3.315 on chromosome 4 at 120 cM, and right amygdala volume (h2=0.6958, SE=0.0754, p=1.43×10−23) had a univariate LOD score of 3.103 in the same location. Moreover, bivariate linkage analysis with left amygdala volume and emotion recognition (rhog=0.24) revealed a bivariate LOD score of 3.282 on chromosome 4 at 122 cM, while bivariate linkage analysis with right amygdala volume and emotion recognition (rhog=0.27) revealed a bivariate LOD score of 3.6812 on chromosome 4 at 122 cM. These results support the idea that the shared genetic influence on amygdala volume and emotion recognition is not lateralized to either the left or right amygdala.
Association Analysis
Association analysis was conducted for all genetic variants under the 1 – LOD score confidence interval of the bivariate linkage peak (defined as 120–124 cM) and a factor score derived from amygdala volume and emotion recognition. In total there were 2,053 SNPs in this region, but after taking LD into account (
53), there were 1,023 effective SNPs, necessitating a Bonferroni-corrected alpha of 5.01×10
−5. One variant met the adjusted-significance level: rs2622497 (p=4.40×10
−5), located within an intron of the gene
PDE5A (phosphodiesterase 5A, cGMP specific). Several other variants met a suggestive level of significance that also fell within
PDE5A in addition to a number of variants downstream of the gene, all of which were in varying degrees of LD with the top-ranked variant (see
Table 1 and
Figure 3). Univariate association analysis for each individual trait for rs2622497 did not reach significance either for amygdala volume (p=1.0×10
−3) or for emotion recognition (p=4.1×10
−3).
When the SNP rs2622497 was included as a covariate in the linkage analysis of the factor score derived from emotion recognition and amygdala volume, the LOD score observed without the covariate (3.336) was reduced (2.506) and no longer significant. This linkage conditional on association test gives additional support for the association between rs2622497, emotion recognition performance, and amygdala volume.
Given the significant effect of age and sex on amygdala volume, the interactive effects of genotype, sex, and age were included as covariates in the association analysis. For our top SNPs, there was a significant interaction with sex (sex by rs2622497: β=0.9823664, p=0.0032331; sex by rs2715021: β=0.9887387, p=0.0032051; sex by rs9884801: β=0.9857738, p=0.0038682; see Figures S1–S3 in the online data supplement) but not with age. In each case, the sex-by-SNP interactions indicated that the effect was marginally more pronounced in men than in women, although the direction of the effect was the same in both groups. Consequently, an interaction term for sex by genotype was included in all association analyses.
Discussion
While numerous studies have highlighted an association between the amygdala and emotion recognition (
10,
19–
22), the present study extends this finding by providing evidence for a pleiotropic locus on chromosome 4 using bivariate linkage. Furthermore, by examining variants within the quantitative locus, we identified a variant (rs2622497) within an intron of
PDE5A that appears to jointly influence amygdala volume and emotion recognition performance. To our knowledge, this is the first study to formally test the common genetic influences on amygdala volume and emotion recognition ability using a bivariate model. It is well established that the implementation of a bivariate as opposed to a univariate model is beneficial, as the joint analysis of multiple traits confers greater power and precision in the mapping of a QTL (
33,
54,
55).
The cyclic nucleotide phosphodiesterases (PDEs) are a family of enzymes with two main subtypes, cAMP- and cGMP-specific nucleotides. The
PDE5A gene codes for PDE5, a cGMP-specific PDE (
56). cGMP, a second messenger, is crucial to signal transduction between cells as well as synapse communication and synaptic plasticity (
57). Hence PDEs are important for effective cell-to-cell communication in the central nervous system (
56). Indeed, there is widespread expression of
PDE5A throughout the body, and it is expressed in various regions of the brain, including in the amygdala (
58), the pyramidal cells of the hippocampus, Purkinje cells in the cerebellum, and some areas of the cortex (
59,
60). Thus, PDE5 has emerged as a potential drug target for treating cognitive deficits (
56,
61). Through the use of mouse models, it has emerged that administration of sildenafil, a PDE5 inhibitor, improves memory, including recognition memory as well as spatial and fear-conditioning memory, in aged rats (
62,
63). Sildenafil also ameliorates cognitive deficits associated with Huntington’s chorea and Alzheimer’s disease by increasing cGMP levels in the hippocampus (
64–
66). Thus the results of the present study are in line with previous research. As in previous studies, the results presented here show an association between recognition memory—of which emotion recognition could be considered a sub-type (
67)—and PDE5.
Recognition is predicated on knowledge retention, which is enhanced by contextual association. In the case of emotion recognition, this might include associations formed by previous experiences in which a particular facial configuration has come to be associated with a particular emotion. Such examples might include life events that precede or co-occur with the expression of a particular emotion, with what a person said while experiencing that emotion or what was said about them during that time, with how one felt on seeing the expression, and so on (
67). If this is the case, then emotion recognition should require access to long-term memory, even when the emotional stimulus is portrayed by a stranger, so it seems plausible that emotion recognition is supported by a distributed neural network that includes those brain regions typically implicated in memory performance in addition to the amygdala (
10,
68). In this context,
PDE5A, a gene expressed particularly in the cerebellum and hippocampus, is especially interesting. Indeed, the cerebellum and the hippocampus, and for that matter the amygdala, have been implicated in long-term memory activation in humans (
69). It is of note that in the present sample there was a significant genetic correlation between amygdala and both cerebellum (rho
g=0.30, p=2.5×10
−3) and hippocampus (rho
g=0.66, p=2.15×10
−14). A tentative hypothesis generated from the results of this study is that emotion recognition might be improved with the administration of a PDE5 inhibitor, which would be in line with some of the research cited above (
62,
63) and would have substantial implications for those individuals for whom emotion recognition proves difficult, such as schizophrenia patients (
2).
Rare variation is a likely source of family-based linkage signals associated with complex traits (
70,
71). Therefore, it is unsurprising that those SNPs showing the strongest association with amygdala volume and emotion recognition in the present study are relatively rare (
Table 1). The use of extended pedigrees, such as those used this study, improves the chances of detecting association with rare variants, as pedigree-based studies represent an implicit enrichment strategy for identifying rare variants. Mendelian transmissions from parents to offspring maximize the chances that multiple copies of rare variants exist in the pedigree. Thus, pedigree-based studies have optimal power to detect effects of rare variants, so it is unlikely that the associations we observed here are false positives, particularly given that the variants in question do not show a significant departure from the Hardy-Weinberg equilibrium (
Table 1). Our top-ranked variant (rs2622497) appears to be similarly rare across populations (
72; see also Table S2 in the online
data supplement).
The results of this study could be called into question if the genetic effects detected were in fact univariate in nature (e.g., driven by only one of the phenotypes, perhaps in particular by amygdala volume). However, the linkage signal was subjected to a pleiotropy test and was shown to be truly pleiotropic. Furthermore, the factor score derived from the factor model can be said to be driven by both traits equally, as the factor loadings, which are used to determine each individual’s factor score, were constrained to be equal. It is true, however, that no formal test can be applied to the association analysis to further underscore the bivariate underpinnings of the signal.
Some evidence from previous research is suggestive of a modulatory effect of age on amygdala volume and emotion recognition (
73–
75) as well as interactions between sex and genotype on amygdala volume (
76). In the present study, the effects of age and sex were controlled for in all analyses, including in the association analysis, where an additional interaction covariate (sex by SNP) was included. A significant sex-by-SNP interaction was evident for amygdala volume for our top three SNPs (see Figures S1–S3 in the online
data supplement), such that the effect of genotype was slightly more pronounced in men than in women, but the direction of effect of was the same.
Patients with schizophrenia exhibit substantial and robust impairments in emotion recognition ability (
39,
77). It is interesting, then, that the gene implicated in this study,
PDE5, codes for a member of the phosphodiesterase enzyme family, as phosphodiesterase genes, and in particular cAMP-specific
PDE4, have been implicated in schizophrenia risk (
78,
79). Moreover, administration of rolipram (a PDE4 inhibitor) reduces phencyclidine-induced cognitive impairments in humans (phencyclidine is an established pharmacological model of schizophrenia symptomatology) (
80). Phosphodiesterases have also been shown to have potential utility in the treatment of Alzheimer’s disease and in particular the associated cognitive impairment (
81,
82). Furthermore, the administration of a PDE5 inhibitor was shown in a recent placebo-controlled study (
83) to reduce symptoms of depression and cognitive impairment. The established role of phosphodiesterases in psychopathology and cognitive impairment in psychiatric illness, taken together with the results of the present study, highlight the potential utility of PDE5 inhibitors in the treatment of emotion recognition impairments in schizophrenia, depression, and Alzheimer’s disease.
In summary, the linkage and association findings presented here highlight a pleiotropic gene, PDE5A, for amygdala volume and emotion recognition ability. To our knowledge, this is the first study to identify a common genetic locus that influences these two traits. Although this study was conducted in healthy individuals, when taken in the context of previous research, which has shown the potential utility of PDE5 inhibitors as cognitive enhancers, it suggests that PDE5A may be an important target for ameliorating emotion recognition deficits in patients suffering from mental or neurodegenerative illness.