Liability to psychiatric disorders such as schizophrenia, bipolar disorder, and unipolar depression is known to have a significant genetic component, which is partially shared between disorders (
1). Polygenic risk scores (PRSs) are a convenient way of summarizing the contribution of disease-associated variants across multiple loci to give individual-specific estimates of risk (
2). In this issue of the
Journal, Musliner et al. (
3) show that PRSs for psychiatric disorders are associated with progression from unipolar depression to bipolar disorder or psychotic disorder in 16,949 people from the iPSYCH2012 Danish population cohort. Specifically, bipolar disorder PRSs were associated with progression to bipolar disorder, and schizophrenia PRSs were associated with progression to psychotic disorder. Association of PRSs with longitudinal progression is novel and has potential clinical utility in early identification of patients likely to need treatments.
Association of PRSs with disease outcomes has three main aims: to predict individuals who will develop the disease, to refine the definition of disease phenotypes, and to highlight genetic mechanisms relevant to disease. The accuracy of a predictor of disease risk is defined by the true positive rate (the probability that it correctly identifies individuals with the disease as being affected) and false positive rate (the probability that it incorrectly identifies individuals without the disease as being affected). These quantities can be calculated for varying values of the threshold used to identify affected individuals and plotted against each other to give the receiver operating characteristic (ROC) curve. The probability that a predictor correctly classifies disease status is measured by the area under the ROC curve, the AUC. The higher the AUC, the more accurate the predictor, with a random predictor having an AUC of 0.5. Typically, a predictor requires an AUC of 0.8 to be regarded as clinically useful (
4). While this criterion has been reached in Alzheimer’s disease (
5), in general, prediction is much less accurate for psychiatric disorders—for example, a PRS typically achieves an AUC of 0.6–0.7 in schizophrenia (
6) and is weaker still for other psychiatric disorders (
7). The accuracy of genetic predictors is known to be limited by the heritability and prevalence of the phenotype being predicted (
8). Therefore, attention has focused increasingly on using genetics to refine phenotype definition, to reduce the clinical heterogeneity typically observed in traditional psychiatric diagnoses, and to define disease subgroups that map more closely onto the underlying biological mechanisms. For example, schizophrenia PRSs can distinguish between schizophrenia, bipolar disorder with mood-incongruent psychotic features, bipolar disorder with mood-congruent psychotic features, and bipolar disorder without psychosis (
9). Schizophrenia PRSs can also distinguish between schizophrenia and other forms of psychosis (
10).
Disease progression can yield genetically informative phenotypes even in small samples (
11) and is therefore a promising avenue for further study. There have been several studies associating PRSs with disease progression in neurodegenerative disorders, for example, predicting cognitive decline in Parkinson’s disease (
12) and progression of mild cognitive impairment to Alzheimer’s disease (
13). There have been fewer genetic studies of disease progression in psychiatric disorders. Significant associations have been shown between psychiatric PRSs and psychiatric, cognitive, and behavioral phenotypes in childhood and adolescence (
14–
16). These studies suggest that PRSs may predict progression from adolescent traits to psychiatric disorders in adulthood. However, none of them performed the longitudinal follow-up necessary for confirmation of this hypothesis. Jonas et al. (
17) reported the results of a 20-year study of a group of first-admission patients with psychosis, in which schizophrenia PRSs were found to predict increased illness severity, along with worse cognition and which individuals will progress from mood disorder with psychosis to a schizophrenia spectrum disorder. This study is interesting because it showed how genetic risk can be related to progression of psychosis longitudinally in patients and can be used to postulate a hypothesis for genetic risk initially predicting cognitive deficits and negative symptoms prior to an eventual diagnosis of nonaffective psychosis. However, given the small sample size (N=249), the results need to be replicated. By contrast, the study by Musliner et al. (
3) in this issue used a large sample to examine the development of progression to bipolar or psychotic disorders in individuals with unipolar depression, thereby widening the range of psychiatric phenotypes beyond psychosis. Interestingly, PRSs for bipolar disorder were associated with progression to bipolar disorder, with PRSs for schizophrenia associated with progression to psychotic disorder, but the combination of a high PRS for bipolar disorder and a high PRS for schizophrenia was associated with progression to affective psychosis. This suggests the potential both for refinement of the phenotype and genotype to increase the specificity of the association. However, PRSs account for a relatively small proportion of phenotypic variance, thus limiting their clinical utility to predict disease progression.
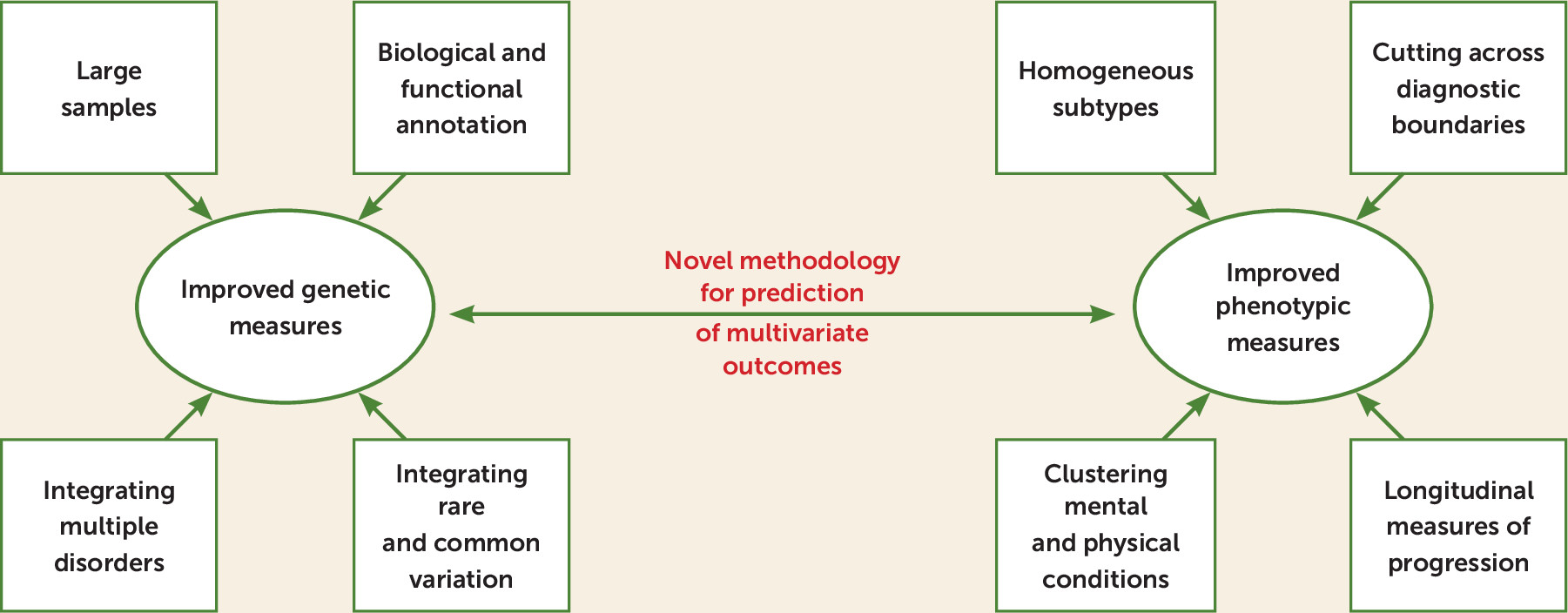
Prospects for the use of genetic risk to predict psychiatric phenotypes center on two main avenues of research: improved genetic measures and improved phenotypic measures (
Figure 1).
Since the predictive ability of PRSs depends on the power of the genome-wide association study (GWAS) used as the training sample (
18), one important way of improving the genetic measures is the collection and analysis of large GWASs of psychiatric disorders. These are currently being coordinated by the Psychiatric Genomics Consortium, and biobanks will also become increasingly important. Methods are also being developed for deriving PRSs that are more powerful than those obtained from the standard p-value thresholding approach (
2), for example, PRSs for continuous shrinkage (
19) and SBayesR (
20). Psychiatric disorders are genetically correlated (
1), such that PRSs from multiple disorders are often associated with clinical phenotypes. Therefore, methods such as genomic structural equation modeling that partition genetic variation from multiple GWASs into portions corresponding to that shared between disorders and disorder-specific components can increase power and specificity of PRS associations (
21). Ultimately, the predictive power of common-variant PRSs is limited by the genetic architecture of the disease (
8). In that case, consideration of rare variants, such as copy number variations (CNVs), may be useful in improving prediction, since the penetrance of these is often high (
22). Furthermore, CNVs associated with neurodevelopmental and psychiatric disorders are associated with psychiatric, cognitive, and behavioral phenotypes in children (
23), suggesting that they may be useful for modeling longitudinal trait progression. There is evidence that an increased number of deleterious rare variants in functionally intolerant genes is associated with reduced IQ in individuals with autism carrying CNVs associated with neurodevelopmental phenotypes (
24) and that schizophrenia PRSs act additively with the 22q11.2 deletion to increase schizophrenia liability (
25). These studies motivate the combination of CNVs with other types of variation to model phenotypic outcomes. The specificity of genetic risk measures for phenotypic prediction may be further improved by incorporating information on biological pathways. This approach has been applied to PRSs in Alzheimer’s disease (
26) and may also be informative in psychiatric disorders. The predictive ability of genetic risk measures may also be improved by integrating expression data (
27) and functional annotation (
28).
It is also crucial to refine definitions of phenotypes to improve the correlation with genetic measures. This can be done simultaneously, for example, by a combination of genomic structural equation modeling and factor analysis to provide gene-phenotype associations that cross traditional disease diagnoses (
29) and thus provide novel biological and clinical insights. As noted earlier, consideration of longitudinal progression is a promising avenue for deriving informative phenotypes with a genetic basis. Intensively phenotyped cohorts are useful in this regard, as they allow multivariate modeling of disease trajectories (
11). Biobanks are another promising source of novel phenotypes. Zemedikun et al. (
30) showed that certain mental and physical conditions clustered together (multimorbidity) in the UK Biobank, although using a cross-sectional, rather than longitudinal, analysis. Recent evidence from the Danish National Registry population cohort indicates that diagnosis of mental disorders influences the future risk of other medical conditions (
31) under a survival analysis (Cox proportional hazards). These studies motivate the construction of clusters of multimorbidity that cut across traditional diagnostic boundaries, and the next step is to discover the genetic (and other) factors that underlie them, ideally in a longitudinal framework.
To conclude, the use of genetics to predict traditional psychiatric outcomes has produced some useful insights into disease mechanisms but has been limited in terms of risk prediction. To address this issue, it will be necessary to derive novel phenotypes that cut across traditional diagnostic boundaries. This will require the analysis of multivariate phenotypic data of various types, including longitudinal progression measures, alongside environmental risk factors. Likewise, genetic predictors should integrate multiple types of variants (common single-nucleotide polymorphisms, rare single-nucleotide variants, and CNVs) with functional and biological information. Analyses of these multidimensional data sets will be challenging and require the development of novel methodology, with machine-learning methods being a promising approach (
32). Criteria for assessing risk prediction in multivariate outcomes (
33) have been proposed to measure the performance of these methods. Thus, while there is still considerable work do be done, the prospects for genomics to accurately predict psychiatric outcomes, and thus target treatments to patients more precisely (
34), are bright.