The Autism Biomarkers Consortium for Clinical Trials: Initial Evaluation of a Battery of Candidate EEG Biomarkers
Abstract
Objective:
Methods:
Results:
Conclusions:
Methods
ABC-CT Protocol
Participant Characteristics
EEG Acquisition Standardization
EEG equipment.
EEG protocol.
Resting-state assay.
Faces assay.
VEP assay.
Biological motion perception assay.
Analytic Plan
Acquisition.
Construct performance.
Six-week stability.
| Measure | Typically Developing | ASD | ASD <8.5 Years Old | ASD ≥8.5 Years Old | ASD IQ ≤75 | ASD IQ >75 |
|---|---|---|---|---|---|---|
| Resting state | ||||||
| Slope | 0.539 | 0.594 | 0.575 | 0.606 | 0.621 | 0.588 |
| Alpha | 0.667 | 0.730 | 0.707 | 0.724 | 0.725 | 0.718 |
| Gamma | 0.453 | 0.555 | 0.561 | 0.452 | 0.476 | 0.542 |
| Faces task | ||||||
| Upright faces P100 latency | 0.687 | 0.680 | 0.799 | 0.592 | 0.895 | 0.667 |
| Upright faces N170 latency | 0.749 | 0.662 | 0.622 | 0.644 | 0.789 | 0.650 |
| Visual evoked potential | ||||||
| VEP N1 amplitude | 0.679 | 0.732 | 0.610 | 0.801 | 0.380 | 0.861 |
| VEP P100 amplitude | 0.743 | 0.700 | 0.752 | 0.570 | 0.820 | 0.693 |
| Biological motion perception task | ||||||
| BMS N200 amplitude | 0.097 | 0.025 | 0.103 | 0.092 | —b | 0.026 |
| BMS P3 amplitude | 0.149 | 0.020 | -0.109 | 0.211 | —b | 0.023 |
Group discrimination.
Phenotypic correlations.
Results
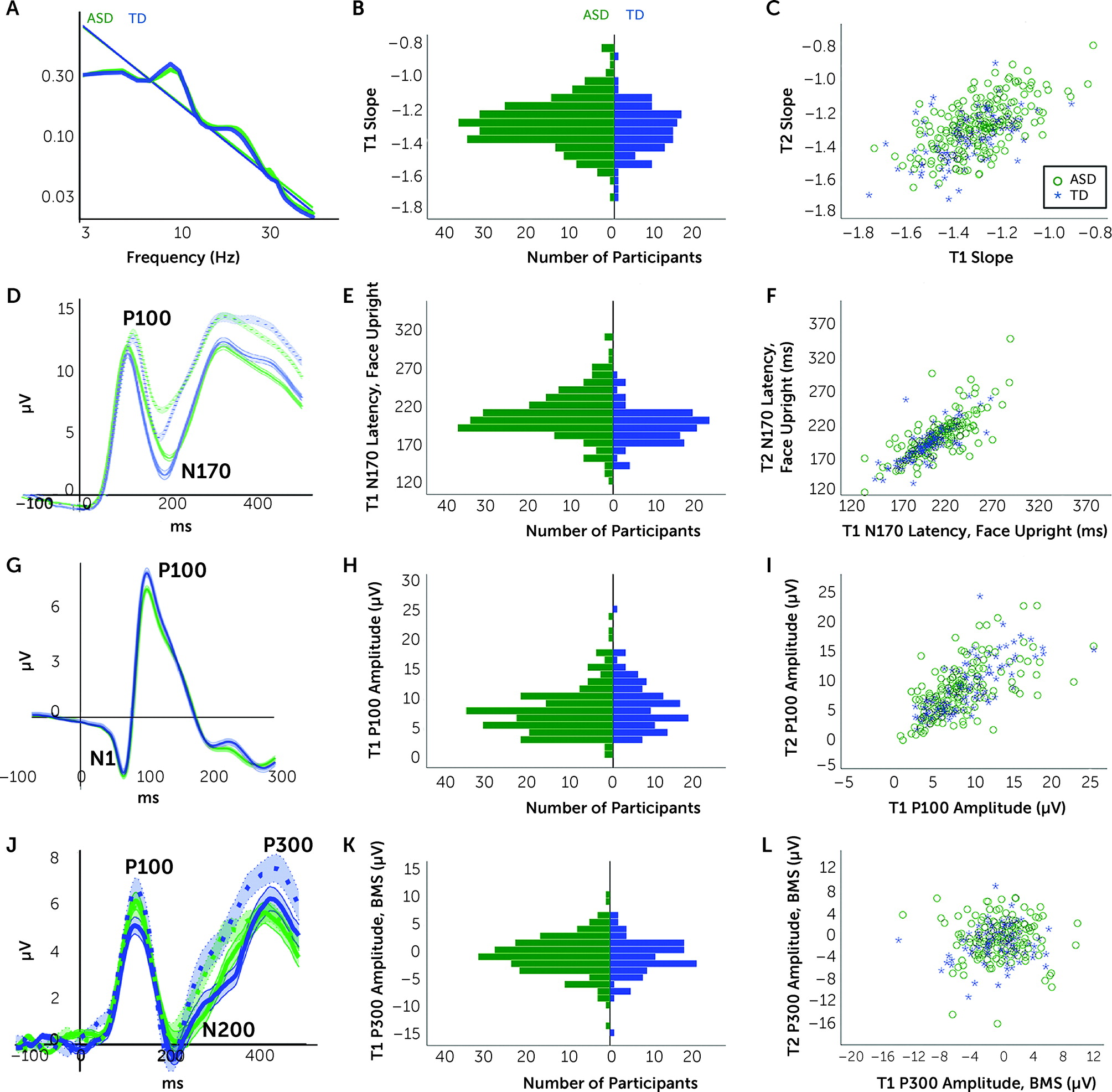
Resting Assay
Acquisition.
| ASD | Typically Developing | Analysis | ||||
|---|---|---|---|---|---|---|
| Measure | Mean | SD | Mean | SD | Analysis Type | Main Effect of Group |
| Resting state | ||||||
| Resting trials | 143.75 | 25.1 | 161.61 | 9.4 | Welch ANOVA | F1,341=93.49, p<0.001, ω2=0.208 |
| Resting slope | −1.279 | 0.144 | −1.317 | 0.139 | ANOVA | F1,350=5.29, p=0.02, ηp2=0.015 |
| ANCOVA | F1,346=1.23, p=0.27, ηp2=0.004 | |||||
| Resting alpha (µV2/Hz) | 0.259 | 0.116 | 0.269 | 0.109 | ANOVA | F1,350=0.57, p=0.45, ηp2=0.002 |
| ANCOVA | F1,346=3.49, p=0.06, ηp2=0.010 | |||||
| Resting gamma (µV2/Hz) | 0.026 | 0.017 | 0.024 | 0.02 | ANOVA | F1,350=0.62, p=0.43, ηp2=0.002 |
| ANCOVA | F1,346=0.44, p=0.51, ηp2=0.001 | |||||
| Faces task | ||||||
| Upright faces trials | 46.38 | 13.9 | 54.31 | 10.5 | Welch ANOVA | F1,295=33.92, p<0.001, ω2=0.091 |
| Upright faces P100 latency (ms) | 121.61 | 17.1 | 117.22 | 12 | Welch ANOVA | F1,307=7.38, p=0.007, ω2=0.012 |
| ANCOVA | F1,324=5.65, p=0.02, ηp2=0.017 | |||||
| Upright faces N170 latency (ms) | 209.63 | 32.4 | 196.86 | 25.7 | Welch ANOVA | F1,284=15.36, p<0.001, ω2=0.042 |
| ANCOVA | F1,324=7.60, p=0.006, ηp2=0.023 | |||||
| Visual evoked potential | ||||||
| VEP trials | 152.52 | 36.5 | 172.83 | 21 | Welch ANOVA | F1,338=43.54, p<0.001, ω2=0.108 |
| VEP N1 amplitude (µV) | −4.39 | 3.3 | −4.53 | 3.5 | ANOVA | F1,331=0.13, p=0.72, ηp2=0.000 |
| ANCOVA | F1,325=0.63, p=0.43, ηp2=0.004 | |||||
| VEP P100 amplitude (µV) | 8.23 | 4.4 | 8.98 | 4.1 | ANOVA | F1,349=2.34, p=0.13, ηp2=0.006 |
| ANCOVA | F1,343=0.39, p=0.53, ηp2=0.001 | |||||
Six-week stability.
Group discrimination.
Phenotype correlations.
| Measure | Trials | Age | Verbal IQ | Nonverbal IQ | Full-Scale IQ | Face Memory |
|---|---|---|---|---|---|---|
| Resting state | ||||||
| Resting slope | −0.261** | −0.177** | −0.110 | −0.050 | −0.080 | −0.163* |
| Resting alpha | −0.214** | −0.261** | −0.147* | −0.146* | −0.171**b | −0.104 |
| Resting gamma | −0.303** | −0.298** | −0.144* | −0.141* | −0.165* | −0.149* |
| Faces task | ||||||
| Upright faces P100 latency | −0.134 | −0.185** | 0.019 | 0.020 | 0.015 | 0.025 |
| Upright faces N170 latency | −0.286** | −0.350** | −0.115 | −0.107 | −0.126 | −0.141* |
| Visual evoked potential | ||||||
| VEP N1 amplitude | 0.109 | 0.241** | 0.093 | 0.013 | 0.049 | 0.123 |
| VEP P100 amplitude | −0.039 | −0.119 | 0.039 | 0.039 | 0.040 | 0.127*b |
Faces Assay
Acquisition.
Construct performance.
Six-week stability.
Group discrimination.
Phenotype correlations.
VEP Assay
Acquisition.
Six-week stability.
Group discrimination.
Phenotype correlations.
Biological Motion Assay
Acquisition.
Construct performance.
Six-week stability.
Group discrimination and phenotype correlations.
Discussion
Acquisition Rates
Construct Performance
Six-Week Stability
Group Discrimination
Phenotype Correlations
Biomarker Viability
Acknowledgments
Footnote
Supplementary Material
- View/Download
- 83.21 KB
- View/Download
- 571.08 KB
References
Information & Authors
Information
Published In
History
Keywords
Authors
Competing Interests
Funding Information
Metrics & Citations
Metrics
Citations
Export Citations
If you have the appropriate software installed, you can download article citation data to the citation manager of your choice. Simply select your manager software from the list below and click Download.
For more information or tips please see 'Downloading to a citation manager' in the Help menu.
View Options
View options
PDF/EPUB
View PDF/EPUBLogin options
Already a subscriber? Access your subscription through your login credentials or your institution for full access to this article.
Personal login Institutional Login Open Athens loginNot a subscriber?
PsychiatryOnline subscription options offer access to the DSM-5-TR® library, books, journals, CME, and patient resources. This all-in-one virtual library provides psychiatrists and mental health professionals with key resources for diagnosis, treatment, research, and professional development.
Need more help? PsychiatryOnline Customer Service may be reached by emailing [email protected] or by calling 800-368-5777 (in the U.S.) or 703-907-7322 (outside the U.S.).

