Approximately one million people die by suicide every year (
1,
2). Thus, it is critical to examine the risk factors associated with suicidality. Several lines of evidence suggest that abnormalities in proinflammatory cytokines may be important contributory risk factors in suicidality (
3,
4). Tumor necrosis factor–alpha (TNF-α), a member of the TNF superfamily, is one of the most important proinflammatory cytokines and plays a critical role in innate and adaptive immune responses (
5). Studies have shown that TNF-α is increased in the serum of depressed individuals who attempt suicide (
6) and that CSF TNF-α levels can serve as a predictor of suicidal ideation (
7). This is supported by another study showing that a composite score of the inflammatory markers, including TNF-α, is associated with high suicidal ideation among patients with major depressive disorder (
8). Early-life adversity, a significant risk factor for suicidal behavior (
9), is also associated with persistent high levels of TNF-α (
10). We previously reported that expression of TNF-α is significantly increased in the prefrontal cortex of adolescents who died by suicide (
11).
Despite evidence of increased TNF-α expression in individuals with suicidal behavior, the underlying regulatory mechanisms are not well understood. Recent evidence suggests the involvement of several genetic and epigenetic mediators in the regulation of TNF-α expression (
12,
13). For example, a genetic association study showed that frequency of the G/G polymorphic locus at position –308 of TNF-α was increased among depressed individuals who died by suicide (
14), as well as among depressed patients who attempted suicide (
15). Furthermore, DNA methylation in the promoter region of TNF-α appears to be crucial in neurodegenerative disorders (
12). An additional layer of epigenetic regulation in TNF-α expression may occur at the posttranscriptional level. This is evidenced by studies showing that microRNAs (miRNAs) are involved in the regulation of proinflammatory cytokines in cancer, age-related macular degeneration, and hemochromatosis (
16–
18). The repressive interaction mediated by miRNAs on target genes is governed by several factors (
19). One of these factors is derived from transactivation response RNA binding protein (TRBP), an miRNA processing protein (
20,
21). TRBP is an integral component of the RNA-induced silencing complex (RISC). One of the functions of TRBP is to transfer the miRNA/miRNA* duplex from Dicer to RISC, which eventually guides miRNA binding to the 3′ untranslated region of target genes (
20,
22). An emerging concept of cis-acting structural RNA motif containing adenylate-uridylate–rich sequence elements on certain transcripts has also gained credence due to regulatory effects (
23). Adenylate-uridylate–rich elements stabilize transcripts by preventing immature decay while interacting with specific sets of RNA-binding proteins, such as Hu antigen R (HuR) (
24,
25). In fact, enhanced expression of TNF-α has been shown to be associated with stabilization of its transcripts containing adenylate-uridylate–rich elements in the 3′ untranslated region mediated by HuR (
26). Endogenous depletion of HuR reverses this stabilization (
27).
Given the multilayer regulation of the TNF-α gene, the present study was undertaken not only to examine TNF-α gene expression but also to understand the precise molecular mechanisms associated with this dysregulation in the brains of individuals who died by suicide. We identified TNF-α expression in the postmortem dorsolateral prefrontal cortex of suicide subjects with major depressive disorder, suicide subjects with other psychiatric disorders, and normal control subjects. The study was also performed in the dorsolateral prefrontal cortex in another cohort consisting of depressed suicide subjects, depressed subjects who died by causes other than suicide, and normal control subjects. The underlying mechanisms of dysregulated TNF-α expression were investigated at genetic and epigenetic levels by examining expression quantitative trait loci, genetic polymorphisms, DNA methylation, miRNAs, TRBP, and HuR. Changes in expression of TNF-α and regulatory miRNA were also examined in a preliminary fashion in the peripheral blood mononuclear cells of suicidal individuals.
Method
Methods are described and discussed in detail in the data supplement accompanying the online version of this article. Additionally, the overall experimental design is presented in Table S1 in the online data supplement. The study was approved by the institutional review board of the University of Alabama at Birmingham.
Human Postmortem Brain Studies
Subjects.
The study was performed in two cohorts: the Quebec Suicide Brain Bank (referred to as the McGill cohort) and the Maryland Brain Collection (referred to as the Maryland cohort). The McGill cohort included the postmortem brains of 16 control subjects without psychiatric disorders (referred to as the normal control group) and 43 brains of subjects who died by suicide (depressed suicide subjects, N=21; suicide subjects with other psychiatric disorders, N=22). The Maryland cohort comprised the brains of 14 depressed subjects who died by suicide, 12 depressed subjects who died by other means, and 12 normal control subjects. Demographic and clinical characteristics of subjects from the McGill and Maryland cohorts are presented in Tables S2 and S3 in the
online data supplement. Psychiatric diagnoses were determined by using the psychological autopsy method, as described in the
data supplement. The study was performed on the dorsolateral prefrontal cortex, since several studies have implicated this brain region in suicidal behavior (
28–
30), and we previously reported abnormalities in the expression of cytokine genes in this region in the brains of individuals who died by suicide (
31).
Relative transcript quantification of TNF-α, TRBP, HuR, miRNAs targeting TNF-α, and members of miR-17-92 cluster.
Total RNA was isolated by using the TRIzol method and screened based on purity (260 nm/280 nm) and RNA integrity number (>7). Since RNA integrity may affect the expression of genes and may pose a confounding factor, we measured the pH and RNA integrity number in all the samples included in this study. The mean pH and mean RNA integrity number are provided in Tables S2 and S3 in the
online data supplement. All subjects had an RNA integrity number >7. Coding gene complementary DNA (cDNA) synthesis was performed with the Oligo (dT)
18 Primer method (Invitrogen, Waltham, Mass.), whereas miRNA-specific cDNA was synthesized by using the poly(A)-tailing method. Relative transcript expressions of all genes were determined with EvaGreen chemistry (Applied Biological Materials, Inc., Richmond, B.C.). The miR-17-92 cluster contains six miRNAs (miR-17, miR-18a, miR-19a, miR-20a, miR-19b-1, and miR-92a-1). The coding units of these six miRNAs produce the following 12 mature miRNAs: miR-17-5p, miR-17-3p, miR-18a-5p, miR-18a-3p, miR-19a-5p, miR-19a-3p, miR-20a-5p, miR-20a-3p, miR-19b-1-5p, miR-19b-1-3p, miR-92-1-5p, and miR-92-1-3p. According to miRBase 21, expression of miR-19a-5p variant is very low (the number of reads is 27 based on deep-sequencing studies). Similar negligible expression of this variant has been observed in human brain, as demonstrated in miRmine (
http://guanlab.ccmb.med.umich.edu/mirmine/), a human miRNA expression database (
32), which appeared to be biologically irrelevant (
33). Therefore, we examined expression of all the miRNAs from this cluster except miR-19a-5p. The primer sequences are presented in Table S4 in the
online data supplement. The results were calculated by using the 2
-ΔΔCt method and reported as fold change.
Genotyping.
A case-control study including 35 control subjects and 60 case subjects was conducted to determine the genetic polymorphism in the TNF-α gene in the genomic DNA isolated from the dorsolateral prefrontal cortex. The TNF-α gene is located on the short arm of chromosome 6 within the major histocompatibility complex, where genetic alterations in the TNF-α locus are directly involved in high TNF-α production (
34). Several polymorphisms have been identified inside the TNF-α promoter (–1031 [T/C], –863 [C/A], –857 [C/A], –851 [C/T], –419 [G/C], –376 [G/A], –308 [G/A], –238 [G/A], –162 [G/A], and –49 [G/A]) (
35). The allelic frequencies of TNF-α have been reported in various populations and are quite consistent (
36), although the polymorphisms at –419, –163, and –49 are rare in Caucasians (
35). Three single-nucleotide polymorphisms (SNPs) (−308G/A: rs1800629, –238G/A: rs361525, and –1031T/C: rs1799964) were selected based on published studies on suicide or depression (
15,
37,
38). Polymerase chain reaction (PCR) amplification products were purified and sequenced with the BigDye Terminator Cycle Sequencing kit (version 3.1; ThermoFisher Scientific, Waltham, Mass.). The primer sequences are summarized in Table S4 in the
online data supplement.
Mutation screening analysis.
Because rare mutations within the seed sequence of miR-19a-3p or the TNF-α 3′ untranslated region may directly affect their canonical bindings, which may lead to altered transcript expression, both the TNF-α 3′ untranslated region and miR-19a-3p were screened for the presence of these mutations. DNA fragments of the TNF-α 3′ untranslated region and miR-19a-3p genes were amplified with PCR using specific primers (see Table S4 in the online data supplement) and sequenced (as described in the Method section in the data supplement).
Methylated DNA immunoprecipitation (MeDIP) assay for TNF-α promoter element.
A potential CpG island was identified within a 1-kb upstream region of the TNF-α transcription start site and analyzed with MeDIP assay by using monoclonal 5-mC antibody. To determine the methylation status, quantitative PCR (qPCR) was used to amplify the CpG island based on primer sequences, as shown in Table S4 in the data supplement, and analyzed after normalization with input control.
Ribonucleoprotein immunoprecipitation assay for detecting HuR interaction with TNF-α adenylate-uridylate–rich elements in the dorsolateral prefrontal cortex.
Relative enrichment of the TNF-α 3′ untranslated region harboring adenylate-uridylate–rich elements was assessed with the ribonucleoprotein immunoprecipitation method, as described in the online data supplement. The primer sequences are discussed in Table S4 in the data supplement.
In Vitro Cell Line–Based Studies
Luciferase reporter assay for detecting miR-19a-3p interaction with TNF-α 3′ untranslated region.
Luciferase reporter assay was performed with pMIR-REPORT (ThermoFisher Scientific, Waltham, Mass.). pMIR [pMIR-REPORT miRNA Expression Reporter Vector, CMV promoter]–TNF-α plasmid, engineered with a 192-base-pair fragment from the 3′ untranslated region of TNF-α containing miR-19a-3p binding site, was cotransfected in human embryonic kidney (HEK)-293 cells with miR-19a-3p mimics or negative controls, pMIR-TNF-α, and pRL-TK (herpes simplex virus thymidine kinase promoter–Renilla luciferase reporter plasmid). Firefly and Renilla luciferase activities were measured after 48 hours.
Cell line–based TNF-α expression studies in response to miR-19a-3p manipulation.
Transient transfections of miR-19a-3p mimic or scramble were performed on SH-SY5Y cells. Total RNAs were isolated and reverse transcribed into first-strand cDNA to determine the relative TNF-α expression after normalization with glyceraldehyde-3-phosphate dehydrogenase.
Cell proliferation assay.
Cell proliferation in transfected SH-SY5Y cells with miRNA oligonucleotide was measured with MTT assay (3-[4, 5-dimethylthiazolyl-2]-2, 5-diphenyltetrazolium bromide). The formazan crystals formed were dissolved in dimethyl sulfoxide, and absorbance was measured at 570 nm.
Transient transfection of HEK-293 cells for HuR overexpression and knockdown.
HuR small-interfering RNA (siRNA), negative control siRNA, and HuR overexpression vector were transfected with lipofectamine 2000. After 48 hours of transfection, expression of TNF-α and HuR was analyzed at the messenger RNA (mRNA) level and the protein level.
Luciferase reporter assay for HuR interaction with TNF-α adenylate-uridylate-rich element.
The pEZX [firefly/Renilla Duo-Luciferase reporter vector]-TNF-3′ untranslated region containing 3′ untranslated region full-length fragments and HuR overexpression vector were cotransfected into HEK-293 cells. Firefly and Renilla luciferase activities were measured after 48 hours.
Ribonucleoprotein immunoprecipitation assay detecting HuR interaction with TNF-α adenylate-uridylate–rich element in vitro.
HEK-293 cell-derived lysates were immunoprecipitated with HuR antibody. Immunocaptured RNA was used to prepare first-strand cDNA by using the Oligo (dT)18 method. HuR-mediated TNF-α 3′ untranslated region enrichment was examined in cDNA following qPCR assay. The primer sequences are presented in Table S4 in the online data supplement.
Gene Expression Studies in Peripheral Blood Mononuclear Cells of Suicidal Patients
In a pilot study, we examined expression of miR-19a-3p and TNF-α in 12 healthy control subjects and 12 currently depressed patients with severe suicidal ideation. The demographic and clinical characteristics of participants are summarized in Table S5 in the
online data supplement. All participants were evaluated with the Mini International Neuropsychiatric Interview (
39) or the Structured Clinical Interview for DSM-IV-TR Axis I Disorders (
40). Depression severity was determined with the Montgomery-Åsberg Depression Rating Scale (MADRS) (
41). Depressed patients with suicidal ideation had a mean score >4 on item 10 of the MADRS. None of the depressed patients were receiving psychotropic drugs prior to blood sampling. Healthy control subjects had no lifetime history of any psychiatric disorder. Venous blood was collected (between 9 a.m. and 11 a.m.) and processed immediately for peripheral blood mononuclear cell isolation. Peripheral blood mononuclear cell pellets were processed for RNA isolation with the TRIzol method. mRNA- and miRNA-specific first-strand cDNA synthesis and PCR amplifications were performed by using the primer sequences described in Table S4 in the
online data supplement.
Statistical Analysis
Data were analyzed with SPSS (version 23; IBM, Armonk, N.Y.). Comparison between normal control subjects and those with suicidal behavior was performed by using independent-sample t tests. Correlations between miRNAs and mRNAs with covariates were determined with Pearson product-moment correlation analyses. Multiple group comparisons were performed with one-way analysis of variance (ANOVA) followed by Bonferroni corrections. We report only data that were normally distributed. For comparisons of genotypic and allelic frequencies and Hardy-Weinberg equilibrium among individuals with suicidal behavior and control subjects, chi-square tests were conducted with SHEsis software (
42). Expression quantitative trait loci analysis, which accounted for sex and age as covariates, was performed with Matrix eQTL software on the R platform. To statistically measure the genotype effect on TNF-α expression, ANOVA was performed by using function “aov” in R. All p values ≤0.05 were considered statistically significant.
Results
TNF-α Expression Studies
TNF-α expression in the dorsolateral prefrontal cortex: effect of suicide and major depressive disorder.
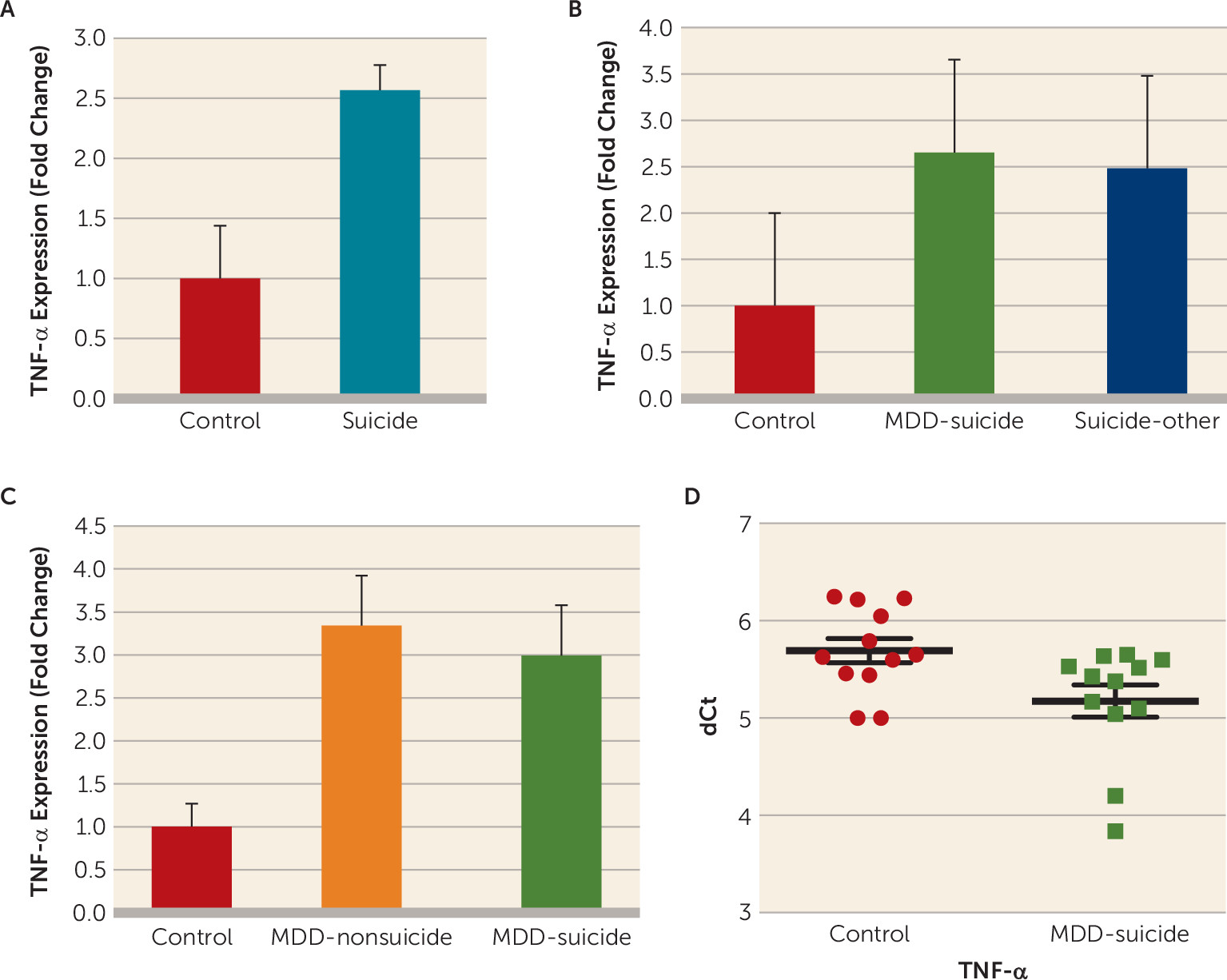
In the McGill postmortem cohort, a statistically significant 2.5-fold greater expression (p=0.004) was found in the dorsolateral prefrontal cortex of the postmortem brains of individuals who died by suicide compared with the postmortem brains of normal control subjects (
Figure 1A). When the suicide group was divided into subjects with major depressive disorder and subjects with other psychiatric disorders, a significant overall difference was observed (F=4.35, df=2, 56, p=0.017). Individual comparisons showed that expression of TNF-α was significantly higher in both depressed suicide subjects (p=0.009) and suicide subjects with other psychiatric disorders (p=0.014), compared with normal control subjects, without any significant difference between the two suicide groups (p=0.83) (
Figure 1B).
To further test whether TNF-α increase was specifically associated with suicide, we examined its expression in the postmortem brains of depressed subjects who died by suicide, depressed subjects without suicidal behavior, and normal control subjects in the Maryland postmortem cohort. Overall, ANOVA showed a significant difference in the expression of TNF-α (F=3.3, df=2, 35, p=0.04). When compared separately, the brains of depressed subjects who died by suicide (p=0.03) and the brains of depressed subjects without suicidal behavior (p=0.027) were significantly different from the brains of normal control subjects, but the two groups of depressed subjects were not significantly different from each other (p=0.83) (
Figure 1C).
In both the McGill and Maryland cohorts, the expression of TNF-α was not related to postmortem interval, brain pH, gender, antidepressant treatment, alcohol use, or substance abuse at the time of death. However, TNF-α showed significant correlation with age in the Maryland cohort but not in the McGill cohort (for further details, see Figures S1 and S2 and Table S6 in the online data supplement).
We combined the data sets from both cohorts and analyzed the expression of TNF-α. A statistically significant (p=0.001; 4.6-fold) increase in TNF-α expression was observed in the brains of suicide subjects (N=57) compared with the brains of normal control subjects (N=28) (for further details, see Figure S3A in the data supplement).
TNF-α expression in peripheral blood mononuclear cells of depressed patients with suicidal behavior.
In the pilot study, expression levels of TNF-α were significantly higher (43%) (p=0.02) in the peripheral blood mononuclear cells of currently depressed patients with severe suicidal ideation compared with healthy control subjects (
Figure 1D), which was not related to age, gender, or race/ethnicity (for further details, see Figure S4 in the
data supplement).
miRNA Expression Studies
Expression analysis of TNF-α targeting miRNAs and miRNA members from miR-17-92 cluster in the dorsolateral prefrontal cortex.
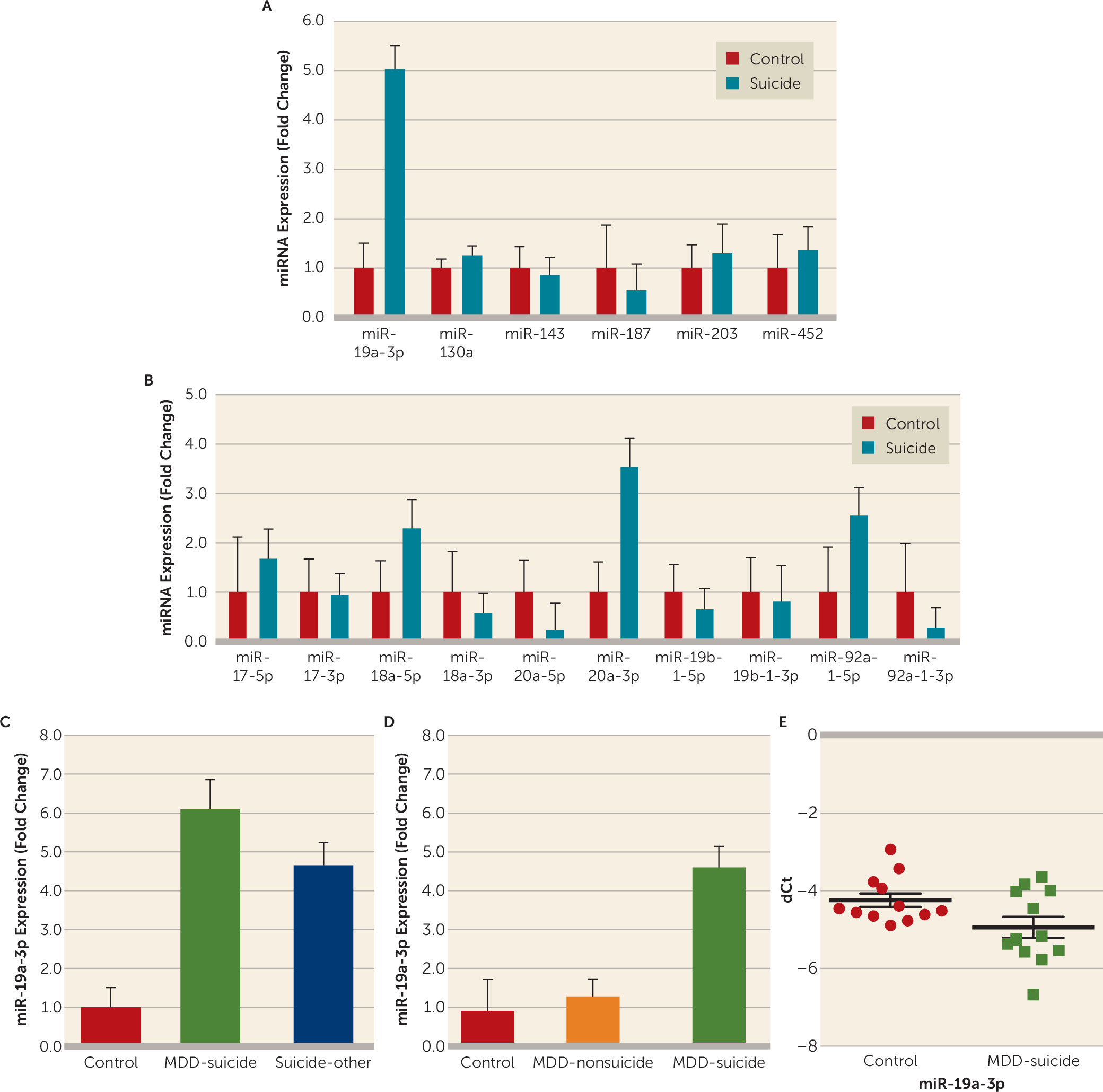
Based on the bioinformatics analysis and target prediction scores, six miRNAs (miR-130a-3p, miR-143-3p, miR-187-3p, miR-203a-3p, miR-452-5p, and miR-19a-3p) were selected as potential repressors of TNF-α expression. qPCR analyses did not show any significant alteration in any miRNA except miR-19a-3p, which was significantly higher (approximately fivefold) among suicide subjects (p=0.008) (
Figure 2A).
Because miR-19a-3p is part of the miR-17-92 cluster, expression levels of 10 other miRNAs (miR-17-5p, miR-17-3p, miR-18a-5p, miR-18a-3p, miR-20a-5p, miR-20a-3p, miR-19b-1-5p, miR-19b-1-3p, miR-92-1-5p, and miR-92-1-3p) associated with this cluster (miR-17-92) were also examined. No significant changes in the expression of any of the cluster miRNAs were observed in the dorsolateral prefrontal cortex of suicide subjects except miR-20a-5p and miR-92a-1-3p, for which expression levels were significantly downregulated (miR-20a-5p: p=0.037; miR-92a-1-3p: p=0.041) by approximately 77% (0.23-fold) and 73% (0.27-fold), respectively (
Figure 2B).
To examine whether observed changes in miR-19a-3p were related to suicide, we separated individuals who died by suicide into two groups: those with major depressive disorder and those with other psychiatric disorders. Overall, a significant group difference was shown with ANOVA (F=3.74, df=2, 56, p=0.03). When examined individually, the expression of miR-19a-3p was significantly higher in both the depressed suicide group (p=0.015; approximately 6.2-fold) and the suicide group with other psychiatric disorders (p=0.024; approximately 4.7-fold) compared with normal control subjects. There was no significant difference between the two suicide groups (p=0.80) (
Figure 2C).
Using the dorsolateral prefrontal cortex from postmortem brains in the Maryland cohort, we examined whether the changes in miR-19a-3p expression were suicide specific. Overall, ANOVA showed significant differences in the expression of miR-19a-3p when the brains of depressed individuals who died by suicide, depressed individuals without suicidal behavior, and normal control subjects were compared with each other (F=3.3, df=2, 35, p=0.04). Individually, miR-19a-3p was significantly higher only in the brains of depressed individuals who died by suicide (p=0.015; approximately 4.6-fold), not in the brains of depressed individuals with no suicidal behavior (p=0.61), compared with normal control subjects. On the other hand, statistical significance was observed when the depressed suicide group was compared with the depressed nonsuicidal group (p=0.04) (
Figure 2D).
The changes in expression of miR-19a-3p in both postmortem cohorts were not associated with postmortem interval, brain pH, and antidepressant toxicology, except that there was a significant correlation with age and miR-19a-3p (r=0.26, p=0.05) in the McGill cohort (for further details, see Figures S1 and S2 and Table S6 in the data supplement).
Combining the data sets from the Maryland and McGill cohorts showed similar significant upregulation (p=0.0001; approximately 4.8-fold) of miR-19a-3p in the dorsolateral prefrontal cortex of individuals who died by suicide compared with normal control subjects (for further details, see Figure S3B in the data supplement).
Expression of miR-19a-3p in peripheral blood mononuclear cells of depressed individuals with suicidal behavior.
When peripheral blood mononuclear cells were examined in the pilot study, the expression of miR-19a-3p was significantly higher in depressed patients with severe suicidal ideation compared with healthy control subjects (63%; p=0.001) (
Figure 2E). The changes in miR-19a-3p expression were not correlated with gender, age, or race/ethnicity (for further details, see Figure S4 in the
data supplement).
In Vitro Validation of TNF-α 3′ Untranslated Region Interaction With miR-19a-3p and Its Effects in the Dorsolateral Prefrontal Cortex of Individuals With Suicidal Behavior
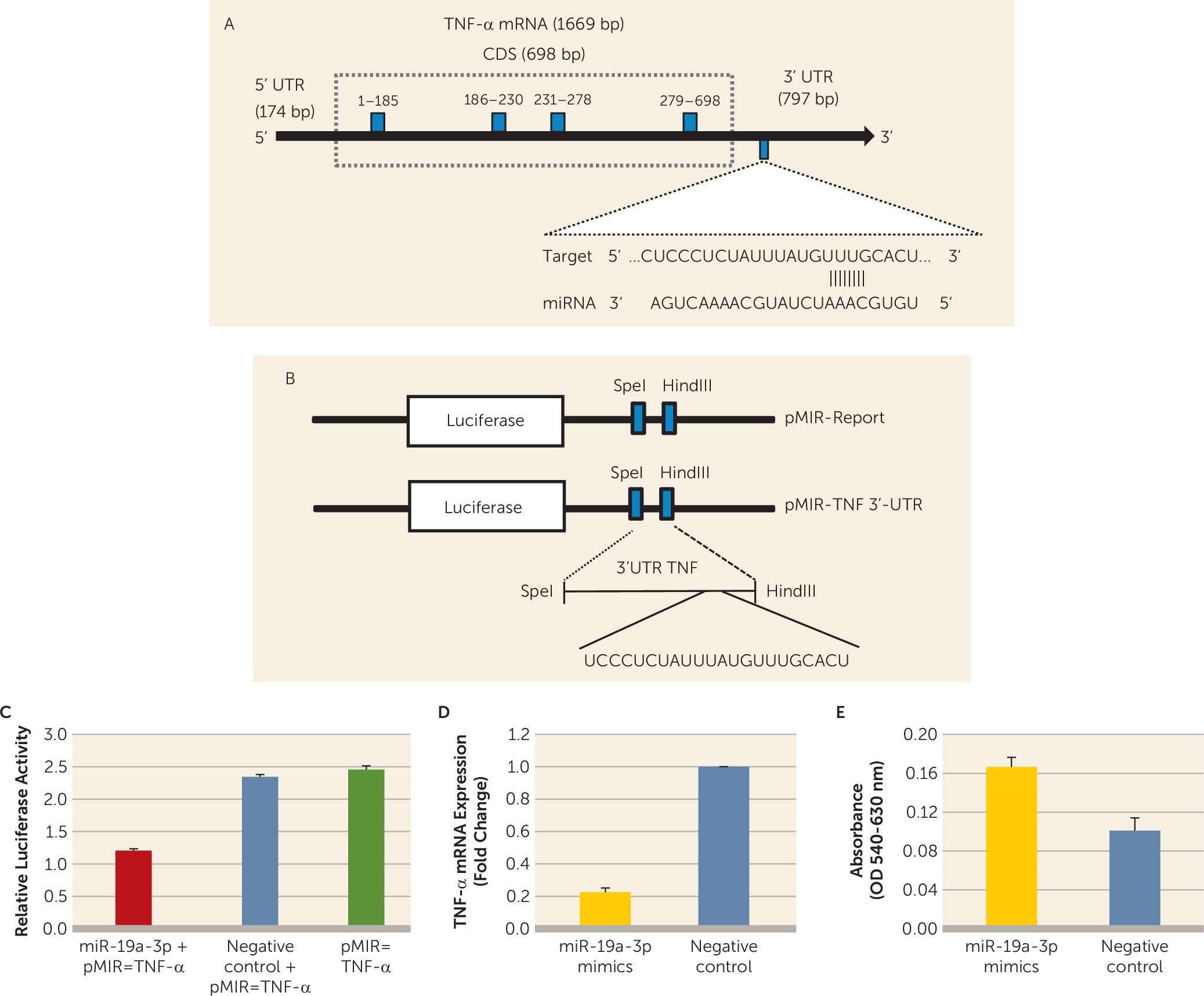
To examine whether miR-19a-3p directly interacts with the TNF-α 3′ untranslated region, HEK-293 cells were transfected with cloned TNF-α 3′ untranslated region downstream of a luciferase reporter gene (
Figures 3A and
3B). Luciferase activity of HEK-293 cells co-transfected with miR-19a-3p mimic was significantly decreased compared with vector alone (approximately 0.49-fold; p=0.0004) or negative controls (0.5-fold; p=0.001) (
Figure 3C). This suggests that TNF-α is a direct target of miR-19a-3p. Under the in vitro condition, functional regulation of miR-19a-3p on the TNF-α gene was determined in SH-SY5Y cells by transfecting miR-19a-3p mimic or negative controls and quantifying TNF-α expression. Transfection of miR-19a-3p significantly decreased the expression of TNF-α (p<0.0001), whereas transfection with negative controls had no effect (
Figure 3D). We next examined whether overexpression of miR-19a-3p had proliferative potential for SH-SY5Y cells. MiR-19a-3p overexpression significantly stimulated cell growth compared with cells transfected with negative controls (p=0.016) (
Figure 3E).
Regulation of TNF-α Expression by TRBP and HuR in the Dorsolateral Prefrontal Cortex
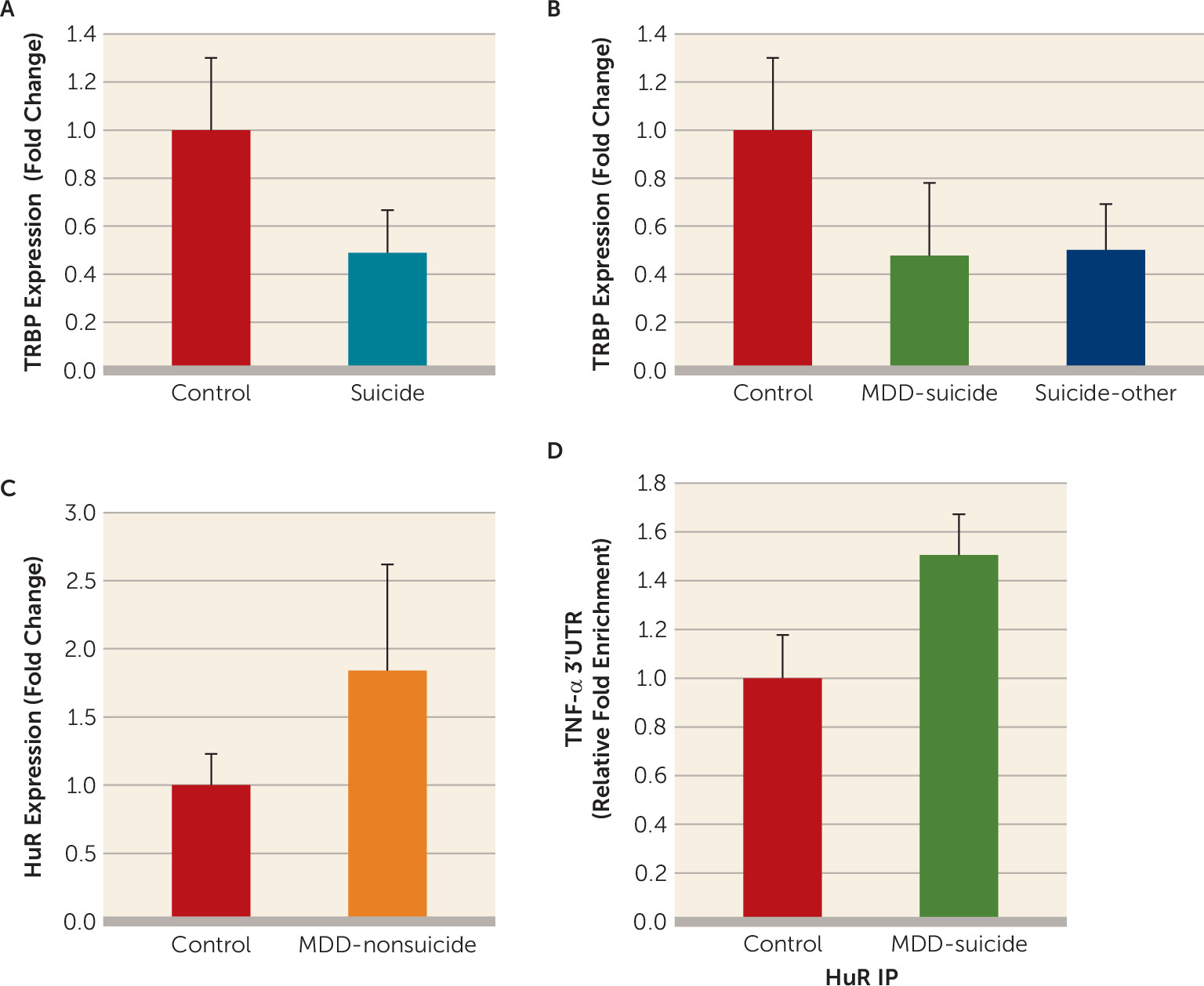
mRNA expression of TRBP was determined in the same RNA fraction in which miR-19a-3p and TNF-α were examined. We observed that expression of TRBP was significantly lower in the suicide group compared with the normal control group (p=0.007; approximately 0.45-fold) (
Figure 4A). This decrease was present in both depressed individuals who died by suicide (p=0.021; approximately 0.44-fold) and individuals with other psychiatric disorders who died by suicide (p=0.012; approximately 0.45-fold) compared with normal control subjects (
Figure 4B).
Next, mRNA expression of HuR was examined in the dorsolateral prefrontal cortex of depressed individuals with nonsuicidal behavior. The expression of HuR was significantly higher (approximately 1.8-fold; p=0.04) in the depressed nonsuicide group compared with the normal control group (
Figure 4C).
In order to examine the role of HuR protein in protecting the TNF-α transcript from repressive effect of miR-19a-3p, endogenous recruitment of HuR on the putative adenylate-uridylate–rich element of the TNF-α 3′ untranslated region was determined with ribonucleoprotein immunoprecipitation assay. Differential binding analysis showed approximately 50% enrichment of HuR protein binding with 3′ untranslated region of TNF-α mRNA in depressed individuals who died by suicide compared with normal control subjects (p=0.028) (
Figure 4D).
To further validate the protecting effect of HuR protein on TNF-α expression, HEK-293 cells were used. HuR mRNA was significantly upregulated (p=0.0008; approximately 15-fold) in the HuR overexpression group compared with the negative control group, whereas HuR mRNA in the siRNA group was significantly decreased (p=0.001; approximately 0.3-fold) when compared with the negative control group (for further details, see Figure S5A in the data supplement). Interestingly, the TNF-α mRNA level in the overexpression group was increased corresponding to the upregulated HuR expression (p=0.005; approximately 14-fold). Contrarily, the TNF-α mRNA level was decreased (p=0.004; approximately 0.6-fold) in the HuR knockdown group due to the downregulation of HuR mRNA. This in vitro assay reveals that TNF-α is positively regulated by HuR (for further details, see Figure S5B in the data supplement).
To further confirm the transient effect of HuR overexpression and knockdown in the HEK-293 cell line, the endogenous protein level of HuR was detected with immunoblotting. The overexpression group showed increased expression of endogenous HuR, whereas the knockdown group showed depleted HuR compared with the nontransfection control group (for further details, see Figure S5C in the data supplement).
Next, in vitro luciferase reporter assay was used to determine the HuR-mediated direct regulation of the TNF-α 3′ untranslated region in the HEK-293 cell line. As shown in Figure S5D in the online data supplement, TNF-α 3′ untranslated region-mediated luciferase reporter activity was significantly increased in the HuR overexpression group compared with the HuR null-transfected group (p=0.0002).
To assess HuR-mediated TNF-α transcript interaction, both ectopically expressed and endogenously depleted HEK-293 cells for HuR protein were used to measure HuR interaction with TNF-α adenylate-uridylate–rich elements. Following the HuR-mediated ribonucleoprotein immunoprecipitation approach, approximately sevenfold enrichment of the TNF-α 3′ untranslated region was observed in the overexpression group compared with the no-transfection control group, whereas a decrease of approximately 50% was observed in the knockdown group (for further details, see Figure S5E in the data supplement).
TNF-α Promoter Methylation in the Dorsolateral Prefrontal Cortex
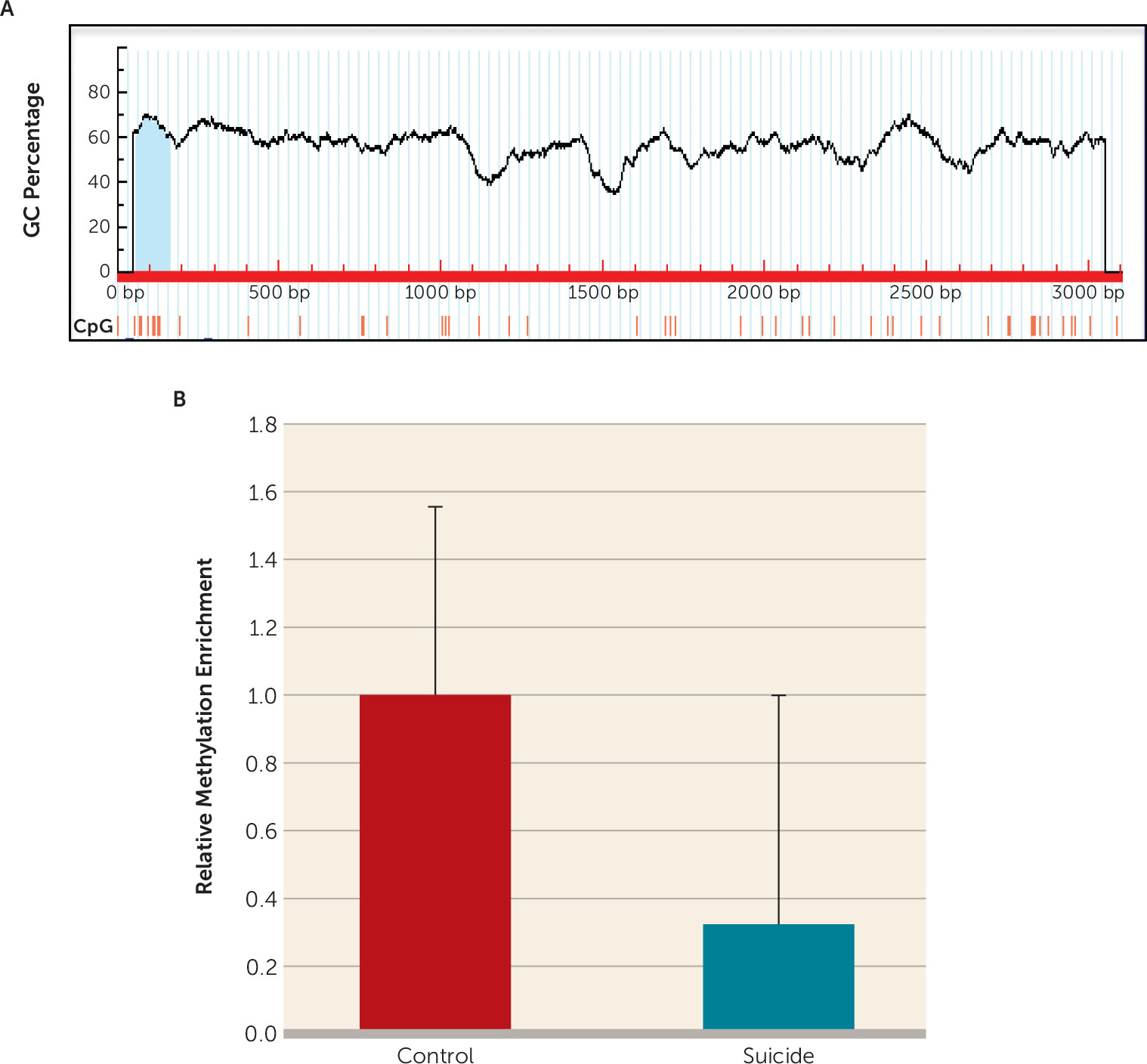
To examine the methylation-associated epigenetic modification as a putative cause of TNF-α upregulation, a CpG island comprising 99 bases and approximately 50% GC contents was identified in the TNF-α gene promoter (
Figure 5A) and analyzed with the MeDIP method. As shown in
Figure 5B, the TNF-α promoter was significantly hypomethylated (approximately 0.7-fold less methylation enrichment) in the suicide group compared with the control group (p=0.039).
Genetic Studies
To examine whether genetic factors were involved in increased TNF-α expression in suicide subjects, three SNPs (rs361525, rs1800629, and rs1799964) in TNF-α gene promoters were studied. Because there was no polymorphism for rs361525 in the TNF-α gene, we excluded this SNP and focused only on rs1800629 and rs1799964. These two SNPs were in Hardy-Weinberg equilibrium. The allelic and genotypic frequencies are presented in Table S7 in the data supplement. There were no significant discrepancies in allelic or genotypic frequencies between individuals who died by suicide and normal control subjects (p>0.05) (for further details, see Table S7 in the data supplement).
To detect the effect of each genotype on mRNA abundance, expression quantitative trait loci analysis was performed. The data revealed no significant difference between the three genotypes for both SNPs (rs1799964, p=0.19; rs18000629, p=0.54) (for further details, see Figure S6 in the data supplement).
To examine whether increased expression levels of miR-19a-3p and TNF-α were associated with duplications or rare mutations, the TNF-α 3′ untranslated region and miR-19a-3p seed DNA sequences were screened for structural variations. As shown in Figures S7A and S7B in the data supplement, no sequence differences for both miR-19-a-3p and TNF-α were observed between suicide subjects and normal control subjects.
Discussion
To our knowledge, this is the first study to examine TNF-α expression regulation in the brains of people who died by suicide. We examined TNF-α expression in two different cohorts of postmortem brain samples and found that expression of TNF-α in the dorsolateral prefrontal cortex was consistently increased in all suicide subjects, regardless of psychiatric diagnosis. A similar change was also observed in depressed individuals who died by causes other than suicide.
Interesting results emerged when we examined the molecular mechanisms associated with increased expression of TNF-α. We first examined the possible regulation of TNF-α by miRNAs, which belong to a novel class of small noncoding RNAs that regulate the gene expression at the posttranscriptional level by binding to the 3′ untranslated region of the target genes guided by RISC. RISC comprises TRBP, Dicer, and argonaute 2 and is functionally responsible for miRNA processing and gene silencing (
22). On the basis of sequence complementarity, prediction score, and an experimentally validated target database, we selected six putative miRNAs (miR-130a-3p, miR-143-3p, miR-187-3p, miR-203a-3p, miR-452-5p, and miR-19a-3p) potentially regulating TNF-α expression. When examined individually, miR-19a-3p was found to be the only miRNA that showed significant upregulation in the dorsolateral prefrontal cortex in those who died by suicide. Interestingly, this upregulation was confined only to those individuals who died by suicide and not observed in those who died by causes other than suicide when compared with normal control subjects. This suggests that change in miR-19a-3p expression in the dorsolateral prefrontal cortex is specific to suicide.
MiR-19a-3p is a member of the miR-19a family, which is encoded by the miR-17-92 cluster. Recently, it has been shown that the miR-17-92 cluster is involved in inflammatory response in malignant human cancers, primarily mediated by miR-19a-3p via TNF-α (
17). Our in vitro luciferase 3′ untranslated region reporter assay in HEK-293 cells and miR-19a-3p mimic transfection assay in neuroblastoma cells ascertain a direct binding of miR-19a-3p with the TNF-α 3′ untranslated region. However, contrary to our in vitro study, we found that the conventional putative interaction was not observed under in vivo conditions, as expression levels of both TNF-α and miR-19a-3p were upregulated in the dorsolateral prefrontal cortex in suicide subjects. This indicates the possibility of sequestration of the TNF-α 3′ untranslated region from the inhibitory effect of miR-19a-3p under in vivo conditions.
MiR-19a-3p belongs to the evolutionarily conserved miR-17-92 cluster, which is located within a locus of the non–protein-coding gene MIR17HG (also referred to as C13orf25) on chromosome 13. In order to test whether upregulation in miR-19a-3p was consistent with other members of the miR-17-92 cluster, we examined the expression of mature miRNAs belonging to this cluster. We found that none of the miRNAs were altered except miR-20a-5p and miR-92-1-3p, which showed significant downregulation. This downregulation was in contrast to the observed change in miR-19a-3p, which was upregulated in suicide subjects. This suggests that miR-19a-3p expression is primarily governed by independent regulatory mechanisms despite being influenced by transcriptional mediators at the miR-17-92 cluster. It is pertinent to mention that mapping of the 3′ untranslated region, even with a less conservative target prediction scheme with TargetScan (version 7.1;
http://www.targetscan.org/vert_71/), did not show any putative binding site for the TNF-α gene for any of the miRNAs belonging to the miR-17-92 cluster except miR-19a-3p (for further details, see Figure S8 in the
online data supplement).
To understand the underlying mechanism of no interaction of miR-19a-3p with TNF-α, we tested whether there were duplications or mutations in the 3′ untranslated region of TNF-α or whether there were mutations in the seed sequences of precursor-miR-19a. Sequencing of the TNF-α 3′ untranslated region revealed no mutations. Additionally, no mutations or duplications were observed in the seed sequences of precursor-miR-19a. These results rule out the possibility that mutations or variations in TNF-α or precursor-miR-19a played a role in preventing the interaction between these two molecules.
Next, we investigated the role of TRBP. We found that expression of TRBP was significantly lower in the dorsolateral prefrontal cortex in suicide subjects. This raises an interesting possibility that less availability of TRBP may be critical in preventing miR-19a-3p binding to TNF-α. It is pertinent to note that we previously reported that pretreatment of rats with a fluoroquinolone compound, which stabilizes the complex between Dicer and TRBP and enhances Dicer-mediated precursor processing and/or loading onto RISC, can ameliorate depressive symptoms (
43), suggesting that aberrant TRBP expression may have phenotypic consequences.
We further tested the possibility that miRNA response may be part of a homeostatic mechanism attempting to pull down TNF-α expression but is thwarted by changes in the accessibility of the TNF-α untranslated region through HuR binding. HuR stabilizes specific transcripts by interacting with adenylate-uridylate–rich elements present in the 3′ untranslated regions of protein coding genes (
44). Interestingly, involvement of target mRNAs of HuR in inflammatory responses has been reported in studies on various disorders (
45). In the present study, HuR antibody-mediated ribonucleoprotein immunoprecipitation assay was used to examine the interaction between HuR and the 3′ untranslated region of TNF-α transcripts in vivo. We observed that this interaction was highly enriched in the dorsolateral prefrontal cortex in suicide subjects compared with normal control subjects, suggesting that increased interaction of HuR with TNF-α could be associated with its increased expression. This was further implied in our study of in vitro HEK-293 cells, in which similar HuR TNF-α transcript interaction was noted under ectopic overexpression of HuR protein, whereas a reduction in HuR-mediated TNF-α transcript interaction was identified in the HuR knockdown model. This mechanism could be critical in preventing the binding of miRNAs to target genes. Thus, HuR can potentially hinder and competitively eliminate miR-19a-3p–mediated repressive effect on TNF-α transcript due to induction of a conformational change in RNA structure and subsequent obstruction of RISC-mediated interaction of miR-19a-3p with TNF-α.
In addition to miRNA and HuR-mediated modifications, we examined other epigenetic and genetic factors for their possible roles in TNF-α upregulation in the brains of suicide subjects. For DNA methylation, we identified a potential CpG island within the TNF-α promoter region and performed MeDIP analysis. We found that the TNF-α gene was hypomethylated in the dorsolateral prefrontal cortex in suicide subjects compared with normal control subjects, suggesting that altered promoter methylation may be involved in upregulation of TNF-α in the brains of suicide subjects. For genetic analysis, a case-control study was designed. Based on the literature, three SNPs (−308G/A: rs1800629; −238G/A: rs361525; and −1031T/C: rs1799964) in the TNF-α promoter were selected for genotyping. No significant differences of allelic and genotypic frequency were found between suicide subjects and normal control subjects, suggesting that genetic polymorphisms may not be critical in TNF-α expression changes in the brains of suicide subjects.
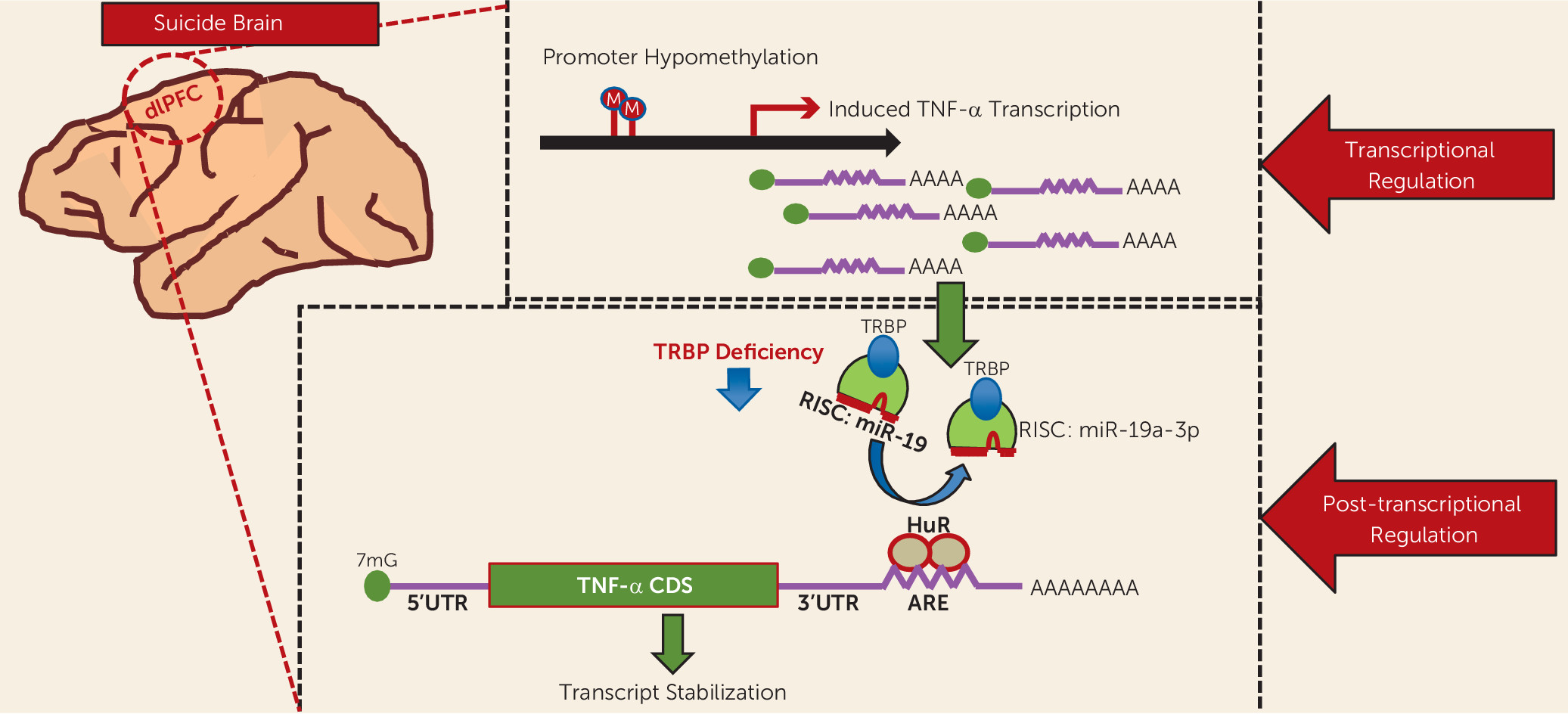
Our study shows not only that TNF-α is significantly upregulated in the dorsolateral prefrontal cortex in the brains of suicide subjects but that there is a complex epigenetic regulatory mechanism that governs this increase. A summary of these results and a putative hypothesis of TNF-α upregulation are presented in
Figure 6. It appears that several molecules, such as miR-19a-3p, TRBP, and HuR, may significantly contribute to this upregulation. HuR may affect TNF-α expression independently of miR-19a-3p. Epigenetic modification mediated by hypomethylation of the promoter region of TNF-α appears to participate in this upregulation. Genetic factors presumably do not play a role in the regulation of TNF-α. Interestingly, in a pilot study (similar to our examination of the dorsolateral prefrontal cortex) we observed that expression of both TNF-α and miR-19a-3p were significantly increased in the peripheral blood mononuclear cells of depressed individuals who had severe suicidal ideation. This indicates that both TNF-α and miR-19a-3p can be examined in blood tissues, which might be associated with suicidality. However, this is a very preliminary observation that has limitations, such that the power of the study is quite low. Furthermore, depressed patients without suicidal ideation were not included in the analyses. Overall, our study provides a novel concept in understanding the molecular basis of altered expression of TNF-α in suicidal behavior.