The psychosis dimension contains complex, disabling mental illnesses, including schizophrenia, schizoaffective disorder, and psychotic bipolar disorder, and evidence indicates significant overlap of clinical features (
1), brain function and structure (
2), pharmacological treatment (
3), and genetic determinants (
4). Psychotic disorders demonstrate high heritability; unaffected family members are at increased risk for all diseases. Studies have implicated shared neurobiological mechanisms of psychosis, including impaired calcium channel activity (
5) and synaptic function (
6). Although copy number variants account for a small proportion of cases (
7), under polygenic models a single gene’s contribution to disease liability is small, but global effects arising from combinations of gene subsets are significant because together they affect specific key neural systems at multiple points (
8). Endophenotypes are heritable, state-independent phenotypes intermediate between the causative genes and clinical disease (
9).
The auditory oddball event-related potential (ERP) is a noninvasively measured electrical brain response elicited by an external auditory stimulus. The P300 is a major ERP subcomponent visible as endogenous positive voltage deflection ∼300 ms after stimulus presentation. P300 amplitude signifies attention allocation, cognitive information processing, and context updating (
10) and is highly heritable (
11). Additionally, auditory ERPs comprise N1, N2, and P2 subcomponents that are generated by the auditory cortices and are associated with stimulus characteristics. Multiple studies report P300 amplitude abnormalities (
12,
13) in both schizophrenia and psychotic bipolar disorder, suggesting general psychosis biomarker status. Auditory P300 amplitude is a frequently employed schizophrenia candidate endophenotype because it is robustly reduced in patients with schizophrenia and their close relatives (
14–
16), is highly heritable, is stable across the course of illness, is relatively unaffected by illness exacerbations, and is elicited by a straightforward task performed easily by psychotic patients. P300 amplitude abnormalities in psychotic bipolar disorder may be proband specific (
17). Deficits in ERP subcomponents N1, P2, and N2 are observed in schizophrenia patients (
18,
19) and their relatives (
20). Both N1 and P2 amplitude abnormalities (
13) and their endophenotypic status have been less studied in psychotic bipolar disorder.
The P300 amplitude response with a parietal maximum (P3b) has been related to norepinephrine (
21) and dopamine neurotransmission (
22). Recent studies examining genetic associations of P300 (
23,
24) adopted traditional single-nucleotide polymorphism (SNP)-based univariate approaches, such as genome-wide association (GWA). One polygenic association study identified several interacting SNP loci, including rs1045642 in the ABCB1 gene, which has been linked to P300 ERP in healthy individuals (
25), plus genes related to dopaminergic, noradrenergic, and signal transduction/amplification pathways. Previous studies (
26–
29) have documented genetic associations of P300 amplitude deficits with known schizophrenia risk genes, including DISC1, NRG1, and COMT. One study (
26) identified rs1045642 as a schizophrenia risk locus.
The univariate GWA approach relates single gene variants to a unitary phenotype. While straightforward, it has two main drawbacks. First, the weak individual effect of common variants prevents most from reaching genome-wide significance, because of numerous multiple comparisons, requiring extremely large samples to gain sufficient statistical power. Second, simultaneous coupling of multiple genes is ignored. To overcome these problems, multivariate association (
30) based on parallel independent component analysis (para-ICA) has been developed to model the polygenic architecture (combinatorial gene effects) of complex psychiatric illnesses. Previous para-ICA studies have revealed known and novel gene clusters implicated in Alzheimer’s disease (
31) and psychosis (
32) based on structural and functional imaging data, respectively.
In the present study, we sought to identify risk genes collectively associated with ERP endophenotypes in psychosis and to determine whether the gene clusters mediate neurophysiological abnormalities in either or both disorders. We hypothesized that para-ICA would identify novel interacting risk gene clusters (likely including known risk genes) mediating plausible physiological pathways that are disturbed in psychosis.
Method
Bipolar-Schizophrenia Network on Intermediate Phenotypes (B-SNIP) Study
We assessed genotype-phenotype multivariate associations by combining genetic variants and ERP from schizophrenia, schizoaffective disorder, and psychotic bipolar disorder probands and healthy comparison subjects from the B-SNIP study (
www.b-snip.org), which was formed to investigate intermediate phenotypes from multiple modalities and their genetic underpinnings in psychosis.
Recruitment
We examined a subset of 620 probands and healthy comparison subjects selected from the overall B-SNIP cohort of ∼2,600 persons (
33) who had undergone genotyping and participated in the B-SNIP auditory oddball paradigm. The group of individuals for whom both genotyping and ERP data were available (N=449) included 144 schizophrenia patients, 210 psychotic bipolar disorder patients, and 95 healthy comparison subjects, ages 15–65 years (demographic and clinical information are provided in Table S1 in the
data supplement that accompanies the online edition of this article). Depressive and mania subtypes of schizoaffective disorder probands were classified as schizophrenia and psychotic bipolar disorder, respectively. Schizophrenia and psychotic bipolar disorder proband groups were not matched with healthy comparison subjects on age, sex, or ethnicity. Probands’ medications are listed in Table S2 in the
data supplement. The study was explained to all participants, and written informed consent was obtained, as approved by the institutional review boards at the participating sites.
Auditory Oddball Paradigm and EEG Data Acquisition
EEG data were collected independently at each site from identical Neuroscan equipment equipped with 64 Ag/AgCl electrodes (Quik-Cap, Compumedics, El Paso, Texas) while subjects were performing an auditory oddball task (see the supplemental text in the data supplement). Electrode positions were defined according to the international 10-10 EEG system (see Figure S1 in the data supplement).
SNP Data Collection
Blood was collected from each subject and genomic DNA was extracted. Genetic variants were obtained by genotyping for ∼1 million (1,140,419) target SNPs across the whole genome using the Illumina Human Omni1-Quad chip and BeadArray platform at Genomas, Inc., at Hartford Hospital.
ERP Data Processing
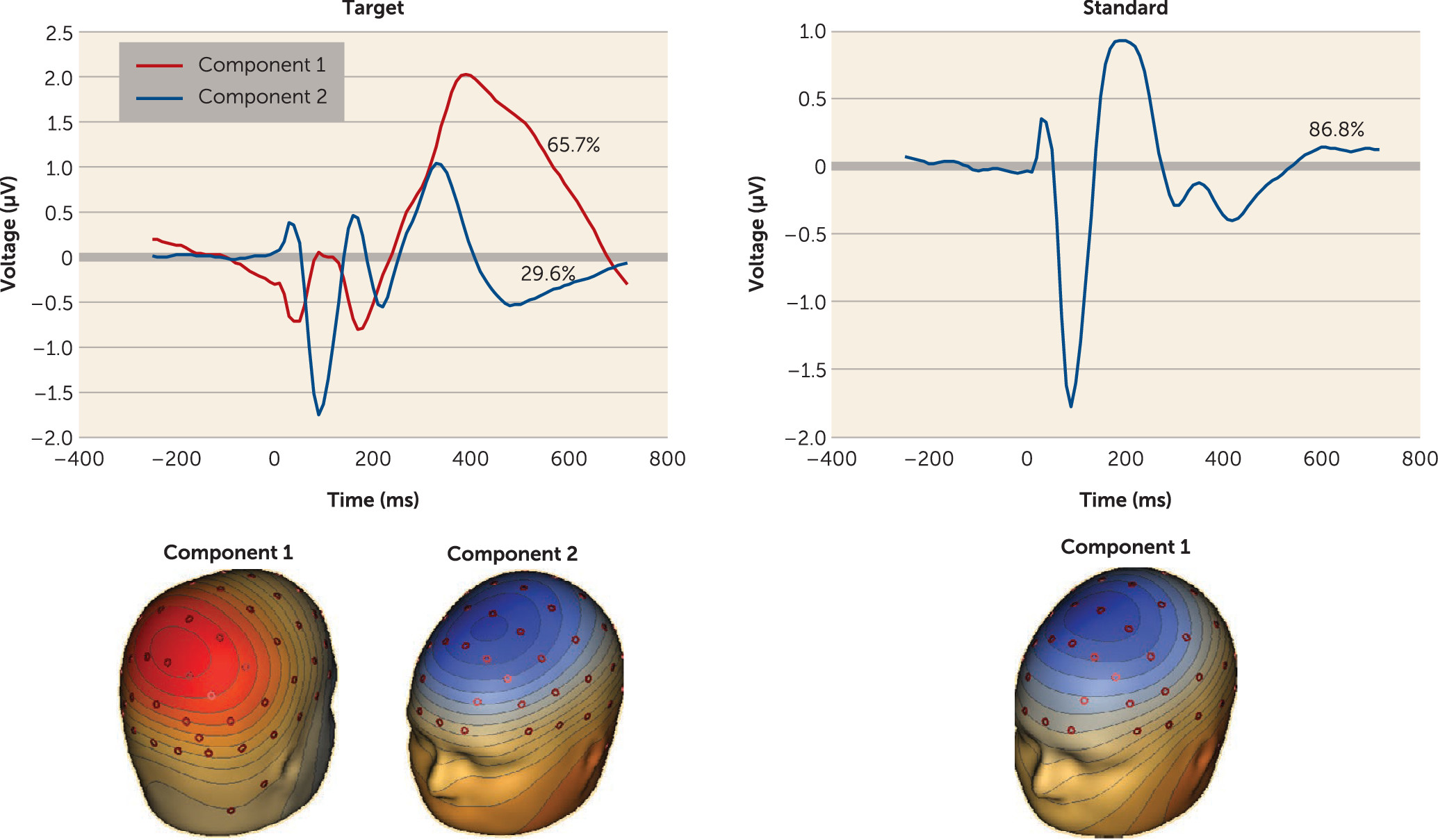
EEG data were artifact rejected, and averaged ERP data were reduced to two targets and one standard component (
Figure 1) using spatial principal components analysis (
13) (details are provided in the
data supplement).
SNP Data Processing
SNP data were converted to discrete numbers and subjected to a quality control process (see Figure S2 in the data supplement). Stratification bias was corrected using principal-components analysis (see Figure S3 in the data supplement). Data were reduced from 1 million to 20,329 SNPs by selecting those that were significant at p<0.05 in the logistic regression (see the data supplement).
Para-ICA-Based SNP-ERP Association
Para-ICA is a multivariate association analysis that links linearly associated functional ERP activity with synergistic gene clusters by relating the hidden structures from both ERP and SNP data. Data were organized as a matrix of 1) subjects by SNPs (449×20,329) and 2) subjects by ERP component waveforms (449×294 [3×98 time points]) and were reduced to 11 and eight components or factors, respectively. ERP and SNP data were jointly processed by para-ICA (
25,
30) (see Figure S4 and the supplementary text in the
data supplement).
Correlation and Statistical Analysis
Bimodal feature association using para-ICA was assessed by correlations between SNP and ERP loading coefficients (LCs). Significance levels were adjusted through partial correlation by regressing the effects of age, sex, and site on LCs. Correlations for all component pair combinations (8×11=88) were evaluated, and significance levels were adjusted using Bonferroni multiple comparison correction for number of combinations (p<0.05/88). ERP and SNP LCs of significant component pairs were examined for group differences using t tests. Dominant SNPs and ERP features in the components contributing to the association were identified with a threshold of |Z|=2.5 for both modalities. Supplementary analyses included correlation of ERP LCs with IQ score and antipsychotic dosage in chlorpromazine equivalents.
ERP-Structural MRI Association Analysis
Brain structures (see the supplementary text for details on structural MRI data collection) associated with the ERP components were identified by partial correlation (accounting for age, sex, and site) of regional volumetric measures with ERP loading coefficients. The p values were corrected for multiple comparisons using false discovery rate.
Pathway Analysis
Genes corresponding to SNPs selected from the genetic component were entered into GeneGo’s MetaCore annotation software package (Thomson Reuters, New York) to determine the biological pathways associated with ERP abnormalities (see the supplementary text).
Results
Gene-ERP Multivariate Linkage
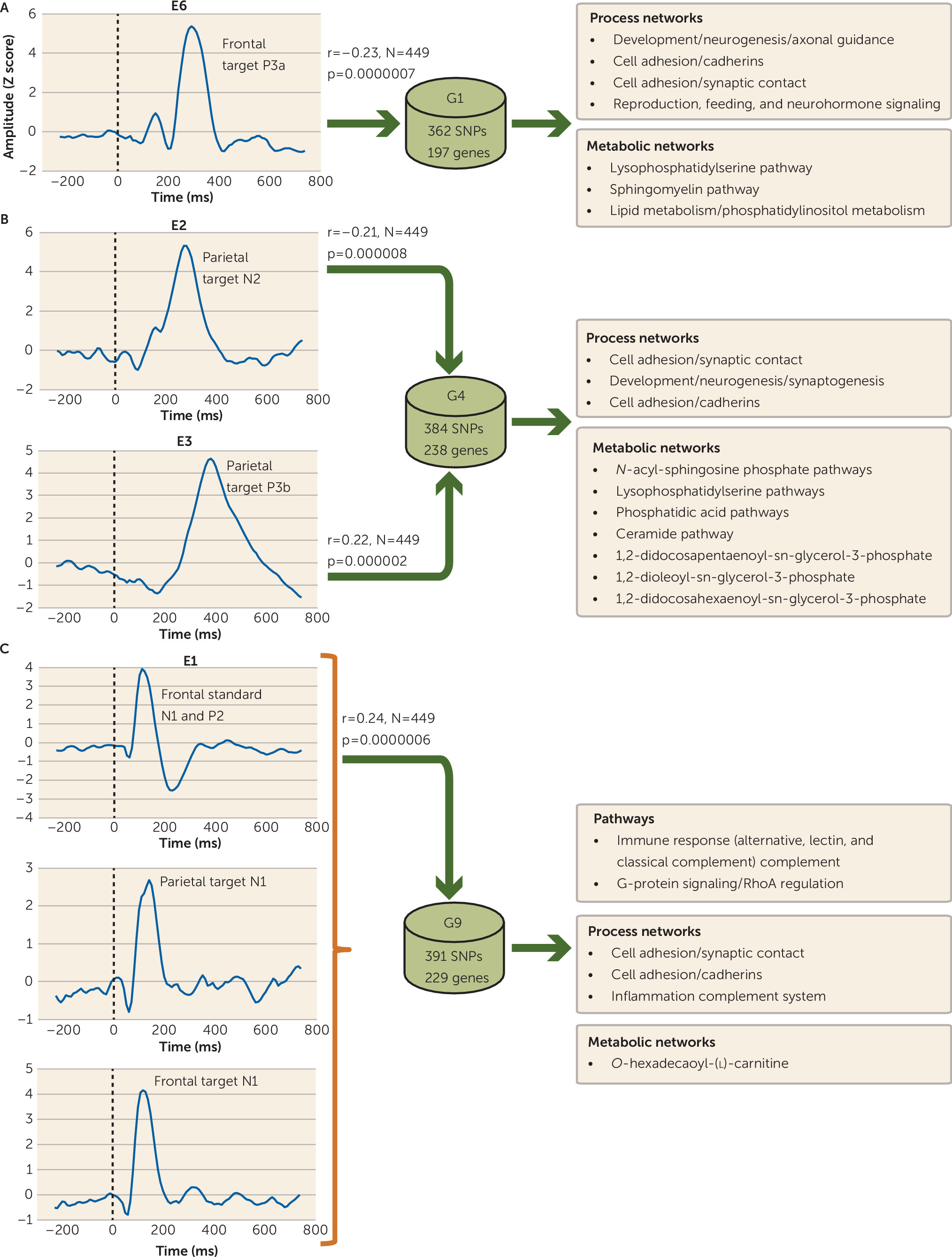
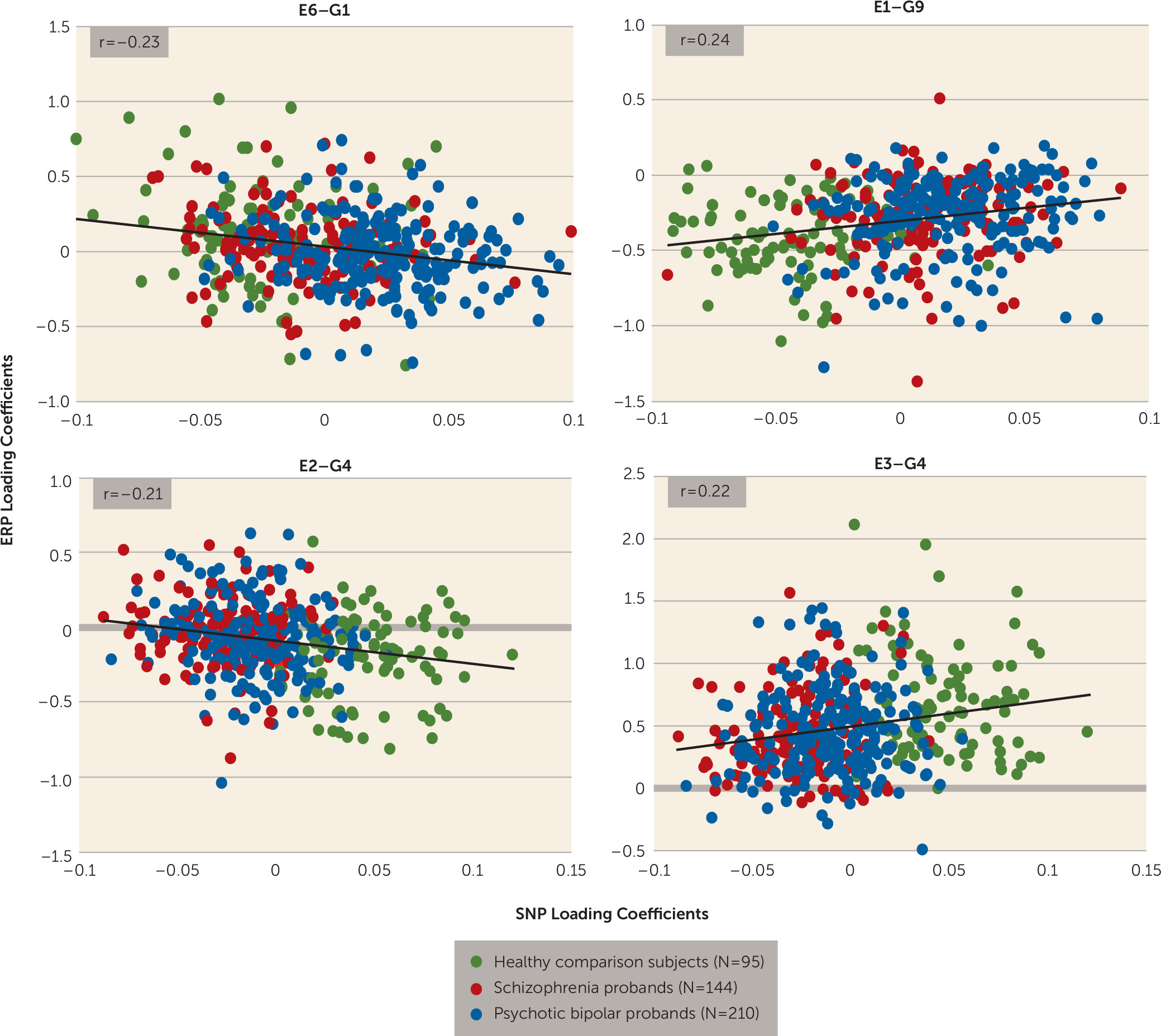
Four (E1, E2, E3, and E6) of the eight independent ERP components were significantly connected to three (G1, G4, and G9) of the 11 genetic/SNP components. ERP component E6 (frontal target P3a) was correlated negatively with G1 (r=−0.23, N=449, p<0.0000007) (
Figure 2A), indicating increased loading on G1 related to diminished frontal target P3a. E1 was positively associated with G9 (r=0.24, N=449, p<0.0000006) (
Figure 2C), indicating increased loading on G9 related to increased ERP features in E1 (frontal standard N1 and P2, and frontal and parietal target N1). Both E2 (parietal target N2) and E3 (parietal target P3b) components were negatively (r=−0.21, N=449, p<0.000008) and positively (r=0.22, N=449, p<0.000002) correlated (
Figure 2B) with the same gene component G4, respectively (
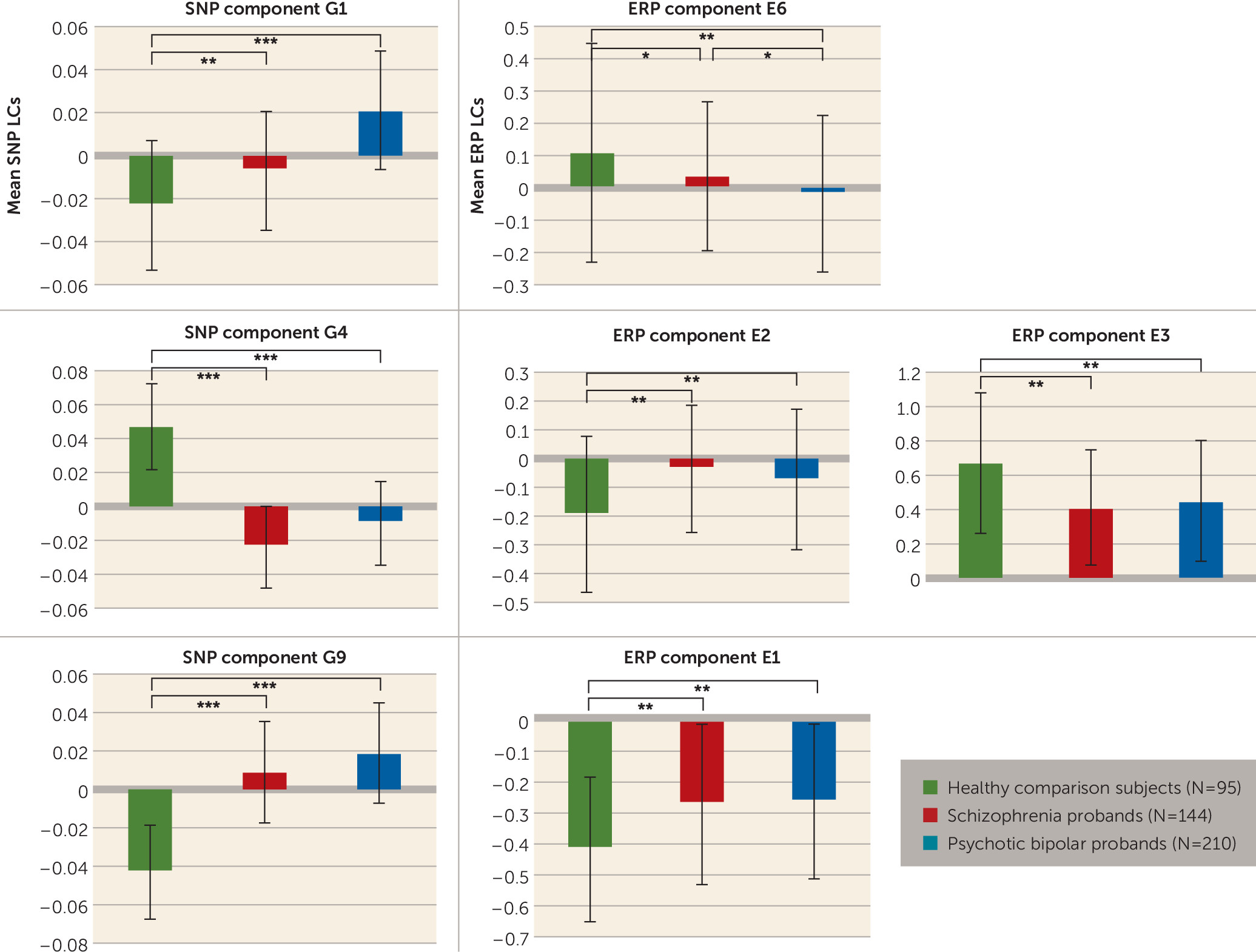
Figure 3). Although IQ differed between groups and was negatively correlated with component E3 (r=−0.166, N=446, p<0.0004), controlling its effects did not alter the ERP-SNP correlation. ERP component E2 was positively correlated with antipsychotic dosage in chlorpromazine equivalents (r=0.179, N=215, p=0.009), indicating that higher dosage was associated with increased abnormality. Post hoc comparisons revealed significant differences in ERP and SNP LCs between healthy comparison subjects and both schizophrenia and psychotic bipolar disorder probands for all significantly correlated components (
Figure 4). ERP component E6 (frontal target P3a) differed between schizophrenia and psychotic bipolar disorder probands, while other ERP components failed to differentiate the two proband groups. Reliability testing by leave-one-out cross-validation revealed an average within-modality correlation >0.85, indicating stable ERP and SNP components.
ERP-Structural Volume Associations
ERP component E1 (frontal standard N1 and P2, and frontal and parietal target N1) was associated with volume reduction in gray matter regions including the pars opercularis, the posterior cingulate, the insula, and the caudal middle frontal, middle temporal, and supramarginal gyri (
Table 1). E3 (P3b) was associated with volume reduction in the left entorhinal and hippocampal regions.
Pathway Analysis
Prominent process networks that were associated with G1 (related to ERP component E6) included (but were not limited to) neurogenesis-axon guidance and cell adhesion (cadherins, synaptic contact) (see Table S3 in the
data supplement). Major metabolic components included lysophosphatidylserine and sphingomyelin pathways. Functional properties for the top 20 genes from all three genetic components are summarized in
Table 2.
Process networks (collection of coordinated genes controlling biological processes) associated with G4 were cell adhesion (synaptic contact, cadherins, and amyloid proteins) and development neurogenesis (synaptogenesis). Several metabolic networks enriched for genes in G4 included (but were not limited to)
N-acyl-sphingosine phosphate, lysophosphatidylserine, phosphatidic acid, and ceramide pathways. Gene component G9 (related to ERP component E1) was significantly (after false discovery rate correction) enriched with immune response genes (classical, alternative, and lectin-induced complements) and G-protein signaling (RhoA regulation). Primary process networks related to G9 were cell adhesion (synaptic contact, cadherins) and the inflammation-complement system. The pathways, known diseases, and major human brain regions from the Allen Brain Atlas (
www.brain-map.org) (assessed based on expression Z scores) that were associated with the top 20 genes are listed in Table S4 in the
data supplement.
Discussion
The etiological mechanisms for schizophrenia and psychotic bipolar disorder are complex, but one approach to gaining insight into them is to examine biological measures such as ERP that are putatively close to the genetic variation. Currently, the neural substrates and genetic basis for eliciting ERP components are unknown. Various ERP subcomponents demonstrate significant heritability, suggesting strong genetic influences. As a preliminary step, we investigated polygenic sources underlying abnormal ERP subcomponents in the psychosis dimension from a large multisite data set including all probands, based on multivariate association. Additionally, we compared schizophrenia and psychotic bipolar disorder probands against healthy comparison subjects based on DSM criteria.
ERP Components
ERP component E1 (a linear combination of standard N1 and P2 and frontal and parietal target N1) showed overall increased loadings (less change from baseline) in schizophrenia and psychotic bipolar disorder compared with healthy comparison subjects, reflecting deficits in early sensory processing, consistent with previous studies in schizophrenia (
18,
20,
34) and psychotic bipolar disorder (
13). The primary source of N1 and P2 is the auditory cortex (
35,
36) (Heschl’s gyrus); we identified reduction in gray matter regions surrounding this structure. Parietal target N2 in E2 was significantly increased (reduced change from baseline) in probands, indexing abnormal stimulus classification (
37). Psychosis probands exhibited decreased frontal target P3a in E6 and parietal target P3b in E3, consistent with previous findings (
12). The role of the hippocampus in P3b generation is debated (
38); however, our finding of P3b amplitude reduction associated with lower hippocampal volume is consistent with previous evidence (
39).
Gene Components
Para-ICA identified gene groups comprising interacting genes with a weight associated with each linkage-contributing gene. We discuss here the genetic underpinnings of ERP abnormalities based on the functionality of top-ranking and frequently appearing genes in each component and biological properties associated with the gene clusters. Of all unique genes pooled across the three genetic components found in this study (N=376), 76 have previously been identified as risk genes for schizophrenia, psychotic bipolar disorder, or both, while we report 300 new genes associated with ERP alterations in psychosis.
G1
Top genes and those identified repeatedly.
The highest-ranking gene in G1 was DCC, encoding a netrin-1 receptor (
40) and a novel schizophrenia candidate risk gene (
41). Multiple intronic SNPs (27 occurrences in the top 50) within DCC were associated with frontal target P3a in E6. The dominant SNP loading was negative, with a negative correlation between G1 and E6, indicating decreased minor allele frequency associated with increased frontal target P3a. DCC plays a critical role in neuronal circuitry, modulating synaptic connectivity by mediating brain development via axon guidance and dendrite growth (
42), with roles in functional reorganization of mesocortical dopamine circuitry (
43). DCC is also involved in the development of brain lateralization (
44), which is abnormal in schizophrenia (
45).
Several intronic SNPs within the BOC gene were found in G1. BOC is the receptor for the molecule Sonic Hedgehog, which plays a critical role in the spatial specificity of synapse formation (
46) and guides the formation of cortical microcircuits (
47) and dorsoventral axon patterning (
48). Multiple SNPs within PDLIM5 encoding the LIM domain protein were detected in G1. PDLIM5 is a candidate risk gene for schizophrenia, psychotic bipolar disorder, and major depression and interacts with brain neuronal
N-type calcium channel and protein kinase-C (
49). Postsynaptic density proteins containing PDZ domain with protein-binding modules interact with glutamate receptors to regulate synaptic plasticity (
50,
51). The HDAC9 gene encoding the histone deacetylase 9 enzyme, which is involved in transcriptional regulation and cell cycle progression, is a known schizophrenia risk gene (
52), is pivotal in neocortical neuron development through transcription regulation via histone modification, and regulates dendritic growth in developing cortical neurons (
53). HDAC inhibitors modulate cell lineage differentiation during brain development (
52) and are direct targets of valproic acid (
54) (a mood stabilizer used to treat psychotic bipolar disorder) and may affect antipsychotic response (
55). Several SNPs within DISC1, a known schizophrenia risk gene, were found in G1 but were not ranked in the top 20.
Pathways, process, and metabolic networks.
Neurogenesis (in particular axon guidance) and cell adhesion (cadherins and synaptic contact) were the enriched functional processes associated with gene clusters in G1 engaged in modulating frontal target P3a abnormality common to psychosis, indicating that these processes are general psychosis risk mediators. Adult neurogenesis likely plays essential roles in both brain function (
56) and the pathophysiology of psychiatric illnesses (
57,
58). The primary neurogenesis-axon guidance-associated genes are PCSK5, CACNA1C, DCC, SLIT3, UNC5C, SLIT1, DISC1, NCAM1, NTRK3, ROBO2, GDA, and PLCB1; of these, several are implicated in psychosis risk.
Another key process identified was cell adhesion mediated by cadherins and synaptic contact. Cadherins depend on calcium ion function to facilitate cell adhesion involved in intracellular signaling, memory formation, neuronal migration, synapse formation, and maturation (
59), associated with neuropsychiatric disorders. Genes associated with cadherin-based cell adhesion were CTNND2, PTPRJ, PCDH15, PDZK3, CTNNA2, CDH12, VAV2, CDH13, and WNT5B. Several of these are implicated in multiple psychiatric disorders; in particular, CDH12 is associated with psychosis. Synaptic cell adhesion molecules mediate neural connections and synapse development. Such genes were SYT9, NRXN3, CTNND2, GRIN2B, CTNNA2, NCAM1, and OBCAM, of which NRXN3 is implicated in schizophrenia and NCAM1 and GRIN2B in both schizophrenia and psychotic bipolar disorder. Sphingomyelin is a signal transduction system that affects neural tissue and neuronal membrane integrity, which suggests that sphingolipid metabolism may play a key role in the pathogenesis of neuropsychiatric disorders (
60).
G4
Top genes and those identified repeatedly.
The most significant G4 candidate gene (associated with parietal target N2 and P3b) was MSRA, a gene that shields against oxidative stress and is associated with schizophrenia susceptibility (
61–
63). Oxidative stress is important in schizophrenia pathophysiology (
64), associated with hypoactive
N-methyl-
d-aspartate glutamate receptors (
65). The chromosome region 8p23.1, in particular SNP D8S542 within MSRA, is strongly associated with both schizophrenia and psychotic bipolar disorder (
66,
67).
Pathways, process, and metabolic networks.
Developmental neurogenesis, synaptogenesis, and cell adhesion was the significant process network mediating psychosis risk through parietal P3b and N2 abnormalities. Metabolic processes including lysophosphatidylserine, phosphatidic acid, and ceramide networks regulate P3b and N2 abnormalities in schizophrenia and psychotic bipolar disorder. Phospholipids constitute cell membranes regulating signal transduction and acting as key secondary messengers. Abnormal phospholipid distribution occurs in schizophrenia in the prefrontal cortex (
68). Both calcium-independent and calcium-dependent phospholipase A2 enzymes are abnormal in schizophrenia (
69,
70). Ceramides are lipid signaling molecules guiding cellular differentiation, proliferation, and apoptosis altered in both first-episode schizophrenia (
71) and depression.
G9
Top genes and those identified repeatedly.
Multiple SNPs were identified in the ME1 gene encoding the cytosolic, NADP-dependent malic enzyme involved in metabolic pathways. No direct evidence from previous studies suggests ME1’s role in schizophrenia and psychotic bipolar disorder, except for increased postmortem expression in the brain in unmedicated patients with bipolar disorder (
72) and linkage signals in schizophrenia in the vicinity of ME1 (
73). The PEMT gene encodes for an enzyme synthesizing membrane phospholipids associated with schizophrenia (
74). This gene helps mediate arachidonic acid signaling, neuronal differentiation, and neurite growth (
75). The GPC6 gene belongs to the glypicans that promote glutamate receptor clustering and receptivity and induce postsynaptic synapse formation (
76). SNAP91 encodes for clathrin coat assembly protein 180, which is enriched in presynaptic neuronal terminals (
77). SNAP91 is involved in intracellular signaling (
78) associated with the Wnt pathway (
79) and is implicated in psychotic bipolar disorder (
80).
Pathways, process, and metabolic networks.
Multiple immune response pathways were associated with psychosis risk mediated by frontal, parietal target N1 and standard N1, and P2 ERP abnormalities in probands, suggesting the immune system’s role in the pathogenesis of psychosis (
81), consistent with previous schizophrenia studies (
82,
83). The classical complement pathway acts on synapse remodeling, pruning, neuronal plasticity, and neurodevelopmental processes via immune system cells and molecules (C3) (
84) and is implicated in the pathophysiology of neurodegenerative disorders (
85,
86). This study implicated immune system CR2 and C3 genes. The latter, a schizophrenia risk gene (
87), is involved in synapse development and regulates neuronal connectivity (
88). Gene component G9 was associated with cell adhesion regulated by cadherins and synaptic contact, which is relevant to schizophrenia and psychotic bipolar disorder risk.
We did not conduct replication analyses to validate the present findings; however, several candidate genes and processes identified in this study have previously been implicated in schizophrenia and psychotic bipolar disorder, supporting the neurodevelopmental hypothesis of schizophrenia (
89) and affective disorders (
90). Our findings point to converging evidence from pathway-based studies that have identified similar biological mechanisms associated with schizophrenia risk, including axon guidance (
91–
93), cell adhesion (
94,
95), inflammation, and the immune system (
96). A recent resting functional MRI-genetic study from B-SNIP using para-ICA (
32) identified similar processes, including developmental neurogenesis, axon guidance, synaptic and cadherin cell adhesion, immune response, nervous system development, and ceramide and sphingosine pathways mediating aberrant default mode activity in psychosis. These data help confirm the present findings and validate the approach undertaken to merge functional and genetic data to dissect the complex mechanisms mediating biological phenotypes in these disorders. A complete overlap in pathways and processes across diverse phenotypes is unlikely, as they probably probe different domains of neurophysiology in these disorders.
Strengths and Limitations
Advantages of this study include our use of high-density spatial ERP data in a large multisite psychosis sample. Limitations included unmatched age, sex, and sample sizes between groups, which we adjusted for by controlling their effects through partial correlation between the SNP and ERP loading coefficients; multisite effects were accounted for by regressing data collection site. The ERP phenotypes we examined may be influenced by medication; it was not straightforward to control for medication effects because probands were on numerous medications and had varying illness chronicity, severity, and duration. Our findings could not be validated because of the lack of a replication sample. Para-ICA was optimized by selecting nominally disease-associated SNPs from univariate analysis. Thus, other genetic networks that may be weakly associated with the ERP subcomponents were overlooked.
Conclusions
As hypothesized, we derived both novel interacting candidate genes and known schizophrenia and psychotic bipolar disorder risk genes mediating ERP abnormalities in psychosis. In general, our data suggest a strong multifactorial genetic component comprising brain-relevant genes that play a key role in neural functions, specifically those controlling neuronal circuits and neurodevelopmental processes. We identified one genetic cluster enriched with genes for neurogenesis-related axon guidance mediating psychosis risk via frontal target P3a abnormalities in probands. The second genetic component was enriched with genes involved in developmental neurogenesis-synaptogenesis, and ceramide and phospholipid networks, influencing P3b abnormalities in psychosis. The third genetic component comprised genes coding for the complement immune response pathway associated with schizophrenia and psychotic bipolar disorder risk through standard N1, P2, and frontal and parietal target N1 abnormalities. All three genetic components mediated ERP abnormalities across the psychosis dimension. Cell adhesion mediated by synaptic contact and cadherins was the prominent network process driving all the ERP abnormalities in psychoses.